| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,041,598 – 7,041,730 |
| Length | 132 |
| Max. P | 0.695707 |

| Location | 7,041,598 – 7,041,730 |
|---|---|
| Length | 132 |
| Sequences | 14 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.57498 |
| G+C content | 0.49441 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.99 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695707 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
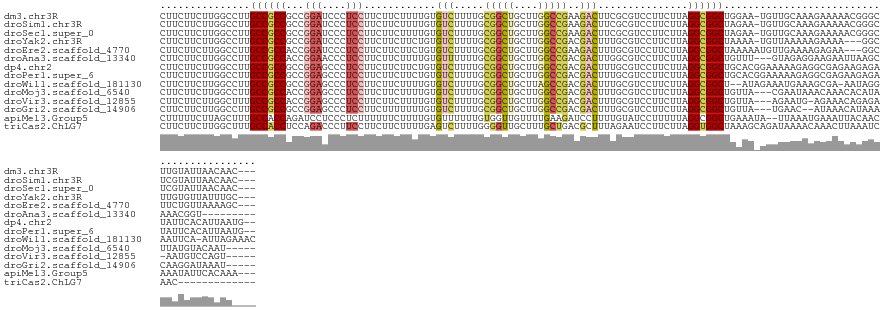
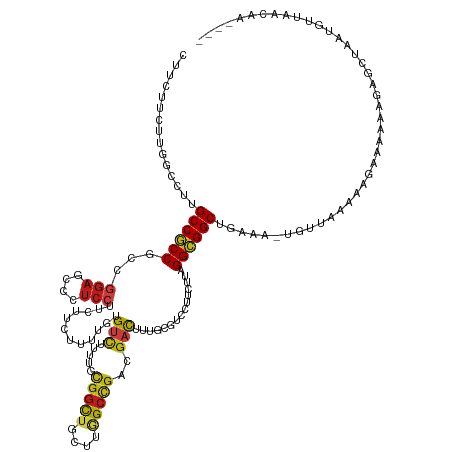
>dm3.chr3R 7041598 132 + 27905053 CUUCUUCUUGGCCUUGCCGCCGCCGGAUCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGAAGACUUCGCGUCCUUCUUAGGCGGCUGGAA-UGUUGCAAAGAAAAACGGGCUUGUAUUAACAAC--- ..(((((..((((..((.(((((.(((..(..............)..)))...))))).))..)))))))))......(((((((((..(((((.....-.))))).)))))....))))((((....)))).--- ( -39.84, z-score = -0.82, R) >droSim1.chr3R 13189307 132 - 27517382 CUUCUUCUUGGCCUUGCCGCCGCCGGAUCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGAAGACUUCGCGUCCUUCUUAGGCGGCUAGAA-UGUUGCAAAGAAAAACGGGCUCGUAUUAACAAC--- ..(((((..((((..((.(((((.(((..(..............)..)))...))))).))..)))))))))...((.(((((((((..(((((.....-.))))).)))))....)))).))..........--- ( -41.24, z-score = -1.40, R) >droSec1.super_0 6203027 132 - 21120651 CUUCUUCUUGGCCUUGCCGCCGCCGGAUCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGAAGACUUCGCGUCCUUCUUAGGCGGCUAGAA-UGUUGCAAAGAAAAACGGGCUCGUAUUAACAAC--- ..(((((..((((..((.(((((.(((..(..............)..)))...))))).))..)))))))))...((.(((((((((..(((((.....-.))))).)))))....)))).))..........--- ( -41.24, z-score = -1.40, R) >droYak2.chr3R 11074006 129 + 28832112 CUUCUUCUUGGCCUUGCCGCCGCCGGAUCCCUCCUUCUUCUUCUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUAAAA-UGUUAAAAAGAAAA---GGCUUGUGUUAUUUGC--- .(((((.(((((.((...(((((((((....)))............(((.....(((((....)))))..)))...............))))))...))-.))))).)))))..---................--- ( -32.60, z-score = 0.01, R) >droEre2.scaffold_4770 3167353 130 - 17746568 CUUCUUCUUGGCCUUGCCGCCACCGGAUCCCUCCUUCUUCUUCUGUGUCUUUUGCGGCUGCUUGGCCGAAGACUUUGCGUCCUUCUUAGGCGGCUAAAAAUGUUGAAAAGAGAA---GGCUUCUGUUAAAAGC--- ..(((((..((((..((.(((...(((....)))..........(((.....)))))).))..)))))))))......(.(((((((...((((.......))))....)))))---))).............--- ( -35.40, z-score = -0.08, R) >droAna3.scaffold_13340 11601820 124 - 23697760 CUUCUUCUUGGCCUUGCCGCCACCGGAACCCUCCUUCUUCUUUUGUGUUUUUUGCGGCUGCUUGGCCGACGACUUGGCGUCCUUCUUAGGCGGCUGUUU---GUAGAGGAAGAAUUAAGCAAACGGU--------- (((((((((((((..((.(((...(((....)))............((.....))))).))..)))))((((...(.((.(((....))))).)...))---))))))))))...............--------- ( -38.00, z-score = -0.29, R) >dp4.chr2 28559618 134 + 30794189 CUUCUUCUUGGCCUUGCCGCCGCCGGAGCCCUCCUUCUUCUUCUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGCACGGAAAAAGAGGCGAGAAGAGAUAUUCACAUUAAUG-- .(((((((((((......)))((((((....))).((((.((((((.......((((((((((((..((((......))))....)))))))))))))))))).)))))))))))))))...............-- ( -49.51, z-score = -2.19, R) >droPer1.super_6 3896615 134 + 6141320 CUUCUUCUUGGCCUUGCCGCCGCCGGAGCCCUCCUUCUUCUUCUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGCACGGAAAAAGAGGCGAGAAGAGAUAUUCACAUUAAUG-- .(((((((((((......)))((((((....))).((((.((((((.......((((((((((((..((((......))))....)))))))))))))))))).)))))))))))))))...............-- ( -49.51, z-score = -2.19, R) >droWil1.scaffold_181130 7794410 132 + 16660200 CUUCUUCUUGGCCUUGCCGCCGCCGGAGCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUAGCCGACGACUUUGCGUCCUUCUUAGGCGGCU--AUAGAAAUGAAAGCGA-AAUAGGAAUUCA-AUUAGAAAC ....((((.(((......)))...(((..(((..(((..((((..(.(((.....((((((((((..((((......))))....))))))))))--..))).)..)))).))-)..)))..))).-...)))).. ( -34.30, z-score = 0.48, R) >droMoj3.scaffold_6540 30422840 128 - 34148556 CUUCUUCUUGGCCUUGCCGCCACCGGAGCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGUUA---CGAAUAAACAAACACAUAUUAUGUACAAU----- ........((((......))))..(((....))).......(((((.((....((((((((((((..((((......))))....))))))))))))..---.))....))))).................----- ( -30.50, z-score = -0.63, R) >droVir3.scaffold_12855 5364240 126 + 10161210 CUUCUUCUUGGCUUUGCCGCCACCGGAGCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGUUA---AGAAUG-AGAAACAGAGA-AAUGUCCAGU----- (((.((((((((...((((((...(((....)))............(((.....(((((....)))))..)))...............)))))).))))---)))).)-)).........-..........----- ( -37.20, z-score = -0.38, R) >droGri2.scaffold_14906 5541096 126 - 14172833 CUUCUUCUUGGCCUUGCCGCCGCCGGAGCCCUCCUUCUUUUUUUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGUUA---UGAAC--AUAAACAUAAACAAGGAUAAAU----- .........(((((.((....)).)).))).(((((.(((.(((((((.....((((((((((((..((((......))))....))))))))))))..---...))--)))))...))).))))).....----- ( -37.30, z-score = -1.69, R) >apiMel3.Group5 5070369 131 - 13386189 CUUUUUCUUAGCUUUGCCACCAGAUCCUCCCUCUUUUUUCUUUUGUGUUUUUUGUGGUUGUUUUGAAGAUCCUUUUGUAUCCUUUUUAGGCGGCUGAAAUA--UUAAAUGAAAUUACAACAAAUAUUCACAAA--- ..(((((..(((...(((((.(((..(...................)..))).))))).)))..)))))....(((((.....((((((....))))))..--......((((((......))).))))))))--- ( -15.41, z-score = 1.32, R) >triCas2.ChLG7 11542464 123 + 17478683 CUUCUUCUUGGCUUUGCCACCUCCAGACCCUUCCUUCUUCUUUUGAGUCUUUUGGGGUUGCUUUGCUGACGCUUUAGAAUCCUUCUUAGGUGGCUAAAGCAGAUAAAACAAACUUAAAUCAAC------------- ..(((..(..((...((.(((((.((((.(..............).))))...))))).))...))..).(((((((...(((....)))...))))))))))....................------------- ( -29.14, z-score = -1.04, R) >consensus CUUCUUCUUGGCCUUGCCGCCGCCGGAGCCCUCCUUCUUCUUUUGUGUCUUUUGCGGCUGCUUGGCCGACGACUUUGCGUCCUUCUUAGGCGGCUGAAA_UGUUAAAAAGAAAAAAGAGCUAAUGUUAACAA____ ...............((((((...(((....)))............(((.....(((((....)))))..)))...............)))))).......................................... (-21.67 = -21.99 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:28 2011