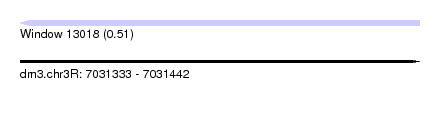
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,031,333 – 7,031,442 |
| Length | 109 |
| Max. P | 0.507546 |

| Location | 7,031,333 – 7,031,442 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.57598 |
| G+C content | 0.53079 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.06 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7031333 109 - 27905053 CAUCCCACAUCUCACAUCUCACAUCUCGGAUCGCAGCC---CAU---CUCACCUGGUCAGCAUCACUUUUUGCAGAUAGGCCUUGCGAUCG-UUUUUGCAGGGCGCCUUGUUGGCU ...(((......................((((((((((---.((---((((...(((.......)))...)).)))).)))..)))))))(-(....)).))).(((.....))). ( -28.60, z-score = -0.77, R) >droWil1.scaffold_181130 7782515 92 - 16660200 ------------------UCACAAUCACAAUCACAA-----CACAAACUCACCUGGUCAACAUCACCUUUUGCAGGUAGGCCUUACGAUCG-UUCUUACAGGGAGCCUUAUUGGCA ------------------..................-----.......((.((((...((((((((((.....)))).(......)))).)-))....))))))(((.....))). ( -16.30, z-score = -0.70, R) >droPer1.super_6 3886085 108 - 6141320 -----GAGCGAAGAGCAGCCACAGCCUGGUCGUCCGAUCUGAUCU--CUCACCUGGUCAGCAUCACCUUCUGGAGGUAGGACUUGCGGUCG-UUCUUGCAGGGUGCCUUAUUGGCA -----((((((...(((((((.....)))).(((((((((((((.--.......)))))).)))(((((...))))).)))).)))..)))-)))........((((.....)))) ( -39.30, z-score = -0.41, R) >dp4.chr2 28549022 108 - 30794189 -----GAGCGAAGAGCAGCCACAGCCUGGUCGUCCGAUCUGAUCU--CUCACCUGGUCAGCAUCACCUUCUGGAGGUAGGACUUGCGGUCG-UUCUUGCAGGGUGCCUUAUUGGCA -----((((((...(((((((.....)))).(((((((((((((.--.......)))))).)))(((((...))))).)))).)))..)))-)))........((((.....)))) ( -39.30, z-score = -0.41, R) >droAna3.scaffold_13340 11592150 97 + 23697760 -----------------AUCUCA-CCAGCUCGACUGGUAGCAUCUCGCUCACCUGGUCAGCAUCACCUUCUGCAGAUAGGCCUUCCGGUCG-UUCUUGCAUGGUGCCUUAUUGGCU -----------------.....(-((((((.((((((((((.....))).))).))))))).........((((((..((((....)))).-.)).))))))))(((.....))). ( -32.60, z-score = -2.18, R) >droEre2.scaffold_4770 3156923 98 + 17746568 --------------CAUCUCUCAUCUCACAUCGCAGCC---CAUCUCCUCACCUGGUCAGCAUCACUUUUUGCAGGUAGGCCUUGCGAUCG-UUUUUGCAGGGCGCCUUGUUGGCU --------------..........((.(((..((.(((---.....((......))...(((........))).)))..(((((((((...-...)))))))))))..))).)).. ( -26.10, z-score = -0.70, R) >droYak2.chr3R 11063814 108 - 28832112 -------CAUCUCACAGCUCACAUCUUACAUCGCAACCCAUCAUCUCCUCACCUGGUCAGCAUCACUUUUUGCAGAUAGGCCUUGCGAUCG-UUUUUGCAGGGCGCCUUGUUGGCU -------.........((((.((....(((((((((((.(((....((......))...(((........))).))).))..))))))).)-)...))..))))(((.....))). ( -26.80, z-score = -1.26, R) >droSec1.super_0 6192670 103 + 21120651 -------CAUCCUACAUCCCACAUCUCGGAUCGCAGCC---CAU---CUCACCUGGUCAGCAUCACUUUUUGCAGAUAUGCCUUGCGAUCGUUUUUUGCAGGGCGCCUUGUUGGCU -------........((((........))))...((((---...---........(((.(((........))).)))..(((((((((.......)))))))))........)))) ( -25.60, z-score = -0.31, R) >droSim1.chr3R 13178942 102 + 27517382 -------CAUCCUACAUCCCACAUCUCGGUUCGCAUCC---CAU---CUCACCUGGUCAGCAUCACUUUUUGCAGAUAUGCCUUGCGAUCG-UUUUUGCAGGGCGCCUUGUUGGCU -------....................((........)---)..---........(((.(((........))).)))..(((((((((...-...)))))))))(((.....))). ( -24.60, z-score = -0.59, R) >consensus _______CAUC__ACAUCUCACAUCUCGGAUCGCAGCC___CAU___CUCACCUGGUCAGCAUCACUUUUUGCAGAUAGGCCUUGCGAUCG_UUUUUGCAGGGCGCCUUGUUGGCU ...............................................((((...(((.......)))...)).))....(((((((((.......)))))))))(((.....))). (-14.93 = -15.06 + 0.12)

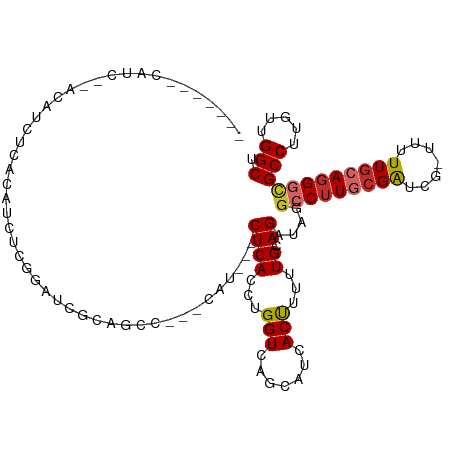
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:27 2011