| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,025,029 – 7,025,129 |
| Length | 100 |
| Max. P | 0.609806 |

| Location | 7,025,029 – 7,025,129 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 59.66 |
| Shannon entropy | 0.74158 |
| G+C content | 0.46246 |
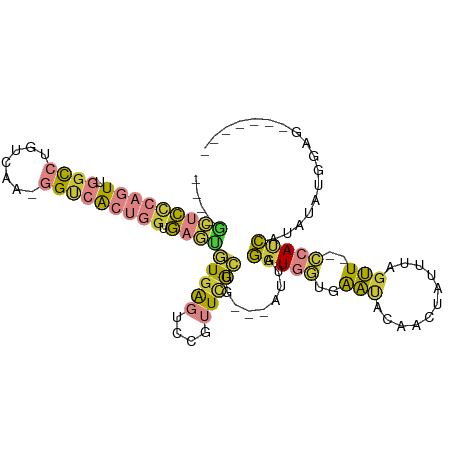
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -9.96 |
| Energy contribution | -10.14 |
| Covariance contribution | 0.18 |
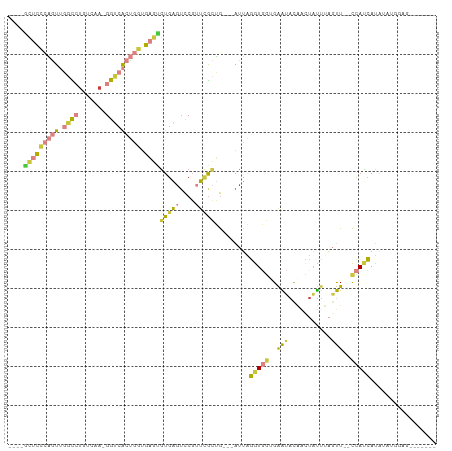
| Combinations/Pair | 1.69 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
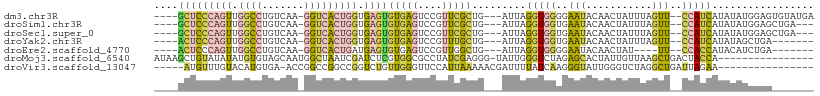
>dm3.chr3R 7025029 100 - 27905053 ----GCUCCCAGUUGGCCUGUCAA-GGUCACUGGUGAGUGUGAGUCCGUUCGCUG---AUUAGGUGGGGAAUACAACUAUUUAGUU--CCAUCAUAUAUGGAGUGUAUGA ----(((((((((.(((((....)-))))))))).)))).......(((.(((((---(((((((((.........))))))))))--((((.....)))))))).))). ( -33.10, z-score = -1.77, R) >droSim1.chr3R 13171709 97 + 27517382 ----GCUCCCAGUUGGCCUGUCAA-GGUCACUGGUGAGUGUGAGUCCGUUCGCUG---AUUAGGUGGUGAAUACAACUAUUUAGUU--CCAUCAUAUAUGGAGCUGA--- ----(((((((((.(((((....)-))))))))).))))...(((..(((((((.---.......)))))))...)))..((((((--((((.....))))))))))--- ( -37.40, z-score = -3.55, R) >droSec1.super_0 6185843 97 + 21120651 ----GCUCCCAGUUGGCCUGUCAA-GGUCACUGGUGAGUGUGAGUCCGUUCGCUG---AUUAGGUGGUGAAUACAACUAUUUAGUU--CCAUCAUAUAUGGAGCUGA--- ----(((((((((.(((((....)-))))))))).))))...(((..(((((((.---.......)))))))...)))..((((((--((((.....))))))))))--- ( -37.40, z-score = -3.55, R) >droYak2.chr3R 11057478 93 - 28832112 ----ACUCCCAGUUGGCCUGUCAA-GGUCACUGGUGAGUGUGAGUCCGUUUGCUG---AUUAGGUGGUGAAUACAACUAUUUAGUU--CCAUCAUAUAGCUGA------- ----(((((((((.(((((....)-))))))))).))))............((((---....(((((((((((....))))))...--)))))...))))...------- ( -28.70, z-score = -1.60, R) >droEre2.scaffold_4770 3150894 89 + 17746568 ----ACUCCCAGUUGGCCUGUCAA-GGUCACUGAUGAGUGUGAGUCCGUUGGCUG---AUUAGGUGGGGAAUACAACUAU----UU--CCACCAUACAUCUGA------- ----((((.((((.(((((....)-))))))))..))))((.((((....)))).---))..(((((..(.........)----..--)))))..........------- ( -27.70, z-score = -0.65, R) >droMoj3.scaffold_6540 21567427 93 - 34148556 AUAAGCUGUAUAUAUGUGUAGCAAUGGCUAAUCGAUCUCGUGGCGCCUAUCGAGGG-UAUUGGGUCUAGAGCACUAUUGUUAAGCUGACUACCA---------------- ...((((..(((...(((((((....)))....(((((....((.(((....))))-)...)))))....))))...)))..))))........---------------- ( -19.20, z-score = 1.75, R) >droVir3.scaffold_13047 14387123 88 + 19223366 -----AUGUUUGUACAUGUGA-ACCGGCCGGCCGGUCUGUUGGGUUCCAUUAAAAACGAUUUUAUCAAGGGUAUUGGGUCUAGGCUGAUUAGAA---------------- -----..(((..(....)..)-))(((((((((.(.((.((((.....(((......)))....)))).))...).))))..))))).......---------------- ( -17.50, z-score = 1.46, R) >consensus ____GCUCCCAGUUGGCCUGUCAA_GGUCACUGGUGAGUGUGAGUCCGUUCGCUG___AUUAGGUGGUGAAUACAACUAUUUAGUU__CCAUCAUAUAUGGAG_______ ....(((((((((.((((.......))))))))).))))(((((....))))).........(((((..(((...........)))..)))))................. ( -9.96 = -10.14 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:24 2011