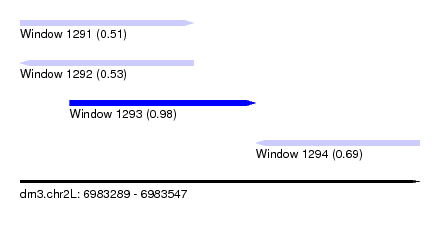
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,983,289 – 6,983,547 |
| Length | 258 |
| Max. P | 0.978204 |

| Location | 6,983,289 – 6,983,401 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.64883 |
| G+C content | 0.52766 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.61 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
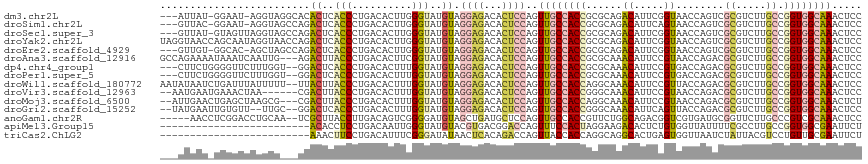
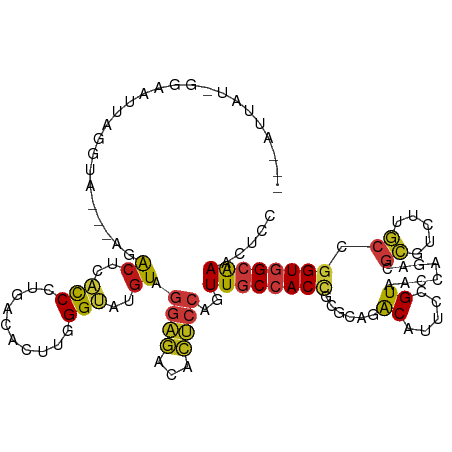
>dm3.chr2L 6983289 112 + 23011544 ---AUUAU-GGAAU-AGGUAGGCACACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCC ---.....-(((..-.((.((....((..((((........))))..)).(....).))))..(((((((((.((((((...(((.......))).))).))))))))))))..))) ( -36.80, z-score = -0.99, R) >droSim1.chr2L 6779177 112 + 22036055 ---GUUAC-GGAAU-AGGUAGCCAGACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCAGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCC ---.....-(((..-..((..((..((..((((........))))..)).))..))..)))..(((((((((.((((((.................))).))))))))))))..... ( -37.23, z-score = -1.39, R) >droSec1.super_3 2516351 113 + 7220098 ---GUUAU-GUAGUUAGGUAGCCAGACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCC ---..(((-(((.((((((.(.(((.......))).))))))).))))))((((...))))..(((((((((.((((((...(((.......))).))).))))))))))))..... ( -37.90, z-score = -1.21, R) >droYak2.chr2L 16398189 117 - 22324452 UAGGUAACCAGCAAUAGGUAACCAGACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCC ..(((.(((.......))).)))..((..((((........))))..)).((((...))))..(((((((((.((((((...(((.......))).))).))))))))))))..... ( -41.40, z-score = -2.24, R) >droEre2.scaffold_4929 15899106 112 + 26641161 ---GUUGU-GGCAC-AGCUAGCCAGACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCC ---....(-(((..-.....)))).((..((((........))))..)).((((...))))..(((((((((.((((((...(((.......))).))).))))))))))))..... ( -39.50, z-score = -1.05, R) >droAna3.scaffold_12916 63101 114 + 16180835 GCCAGAAAAUAAAUCAAUUG---AGACUUACCCUGACACUUCGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCC ...................(---((......((((..((....))...))))...........(((((((((.((((((....(((......))).)).))))))))))))).))). ( -30.80, z-score = -1.92, R) >dp4.chr4_group1 1522760 112 - 5278887 ---CUUCUGGGGUUCUUUGGU--GGACUCACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUGACCAGACGCGUCUUGCCGGUGGCAAACUCC ---...(((((((((((.(((--(....))))(((..((....))...))))))).)))))))(((((((((.((((......((((......))))..)))))))))))))..... ( -39.90, z-score = -1.50, R) >droPer1.super_5 1519940 112 + 6813705 ---CUUCUGGGGUUCUUUGGU--GGACUCACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUGACCAGACGCGUCUUGCCGGUGGCAAACUCC ---...(((((((((((.(((--(....))))(((..((....))...))))))).)))))))(((((((((.((((......((((......))))..)))))))))))))..... ( -39.90, z-score = -1.50, R) >droWil1.scaffold_180772 1645235 115 - 8906247 AAUAUAAUCUGAUUUAUUUUU--UUACUUACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCAGGCAAACAUUCCGUUACCAGACGCGUCUUGCCGGUGGCAAACUCC ........(((......((((--((((.((((..........)))).)))))))).....)))((((((((.(((((((....((((....)))).)).)))))))))))))..... ( -31.80, z-score = -2.85, R) >droVir3.scaffold_12963 3423709 109 + 20206255 --AAUGAAUGAAACUAA------CGACUUACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGGGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCC --...............------..((.((((..........)))).)).((((...))))..((((((((((.(((((....(((......))).)).)))))))))))))..... ( -28.70, z-score = -0.87, R) >droMoj3.scaffold_6500 13797588 112 + 32352404 --AUUGAACUGAGCUAAGCG---CGACUUACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCAGGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCU --.(((.((((.((.(((((---((.(.......)....(((((((((..((((...))))..(((((....)))))......))).)))))))))).)))))))))..)))..... ( -31.10, z-score = -0.72, R) >droGri2.scaffold_15252 2987635 111 + 17193109 --UAUGAAUUGUGUU--UUGC--GGACUCACCCUGACACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGGGCAAACAUUCAGUUACCAGACGCGUCUUGCCGGUGGCAAACUCC --........(((((--..(.--((.....))).))))).((((...((.(....))).))))((((((((((.(((((.....(((....)))..)).)))))))))))))..... ( -31.80, z-score = -0.27, R) >anoGam1.chr2R 36295478 110 - 62725911 -----AACCUCGGACCUGCAA--UCGCUUACCUUGACAGUCGGGGAUGUAGCUGAUGCUCCAGUUGCCACCGUUCUGGCAGACGGUCGUGAUGCGGUUCUUGCCCGUCGCAAACUCC -----.......((((.(((.--(((((..(((((.....)))))....))).)))))....(((((((......))))).))))))((((((.(((....)))))))))....... ( -36.30, z-score = -0.48, R) >apiMel3.Group15 4259502 92 + 7856270 -------------------------ACACCUCCUGACAAUUGGGUAUGUACGUGACGGACCAGUUUCCACUAGGAAGACACUCUGUGGUUAUUUUCGCCUUGCCGGUGGCGAAUUCU -------------------------(((..(((........)))..)))...(.(((((....(((((....)))))....))))).).....((((((........)))))).... ( -21.90, z-score = 0.72, R) >triCas2.ChLG2 10851822 92 - 12900155 -------------------------AAACUUCCUGACAUUUCGGGAUAUAACUCACAGACCAGUUACCACCAGGCAGGCACUGAGUGGUUAAUCUAUUACGUCCUGUUGCGAAUUCU -------------------------.....((((((....))))))........((((((.((..((((((((.......))).)))))....)).....)).)))).......... ( -18.70, z-score = 0.07, R) >consensus ___AUUAU_GGAAUUAGGUA___AGACUCACCCUGACACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCC .........................((..(((..........)))..)).((((...))))..((((((((......((.....))........((.....)).))))))))..... (-13.43 = -13.61 + 0.19)
| Location | 6,983,289 – 6,983,401 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.64883 |
| G+C content | 0.52766 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -15.39 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6983289 112 - 23011544 GGAGUUUGCCACCGGCAAGACGCGACUGGUUACCGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUGUGCCUACCU-AUUCC-AUAAU--- (((((((((((((((((.((((....(((...)))..))))))).)))))))))..((((...))))(((.(((((........))))).))).........-)))))-.....--- ( -39.30, z-score = -1.17, R) >droSim1.chr2L 6779177 112 - 22036055 GGAGUUUGCCACCGGCAAGACGCGACUGGUUACUGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUCUGGCUACCU-AUUCC-GUAAC--- ..((((.((((((((((.((((.....(....)....))))))).)))))))))))(((((((..((.((.(((((........))))).))..))..))..-)))))-.....--- ( -38.70, z-score = -0.70, R) >droSec1.super_3 2516351 113 - 7220098 GGAGUUUGCCACCGGCAAGACGCGACUGGUUACCGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUCUGGCUACCUAACUAC-AUAAC--- (((((((((((((((((.((((....(((...)))..))))))).))))))))(((((((...))))....(((((........))))))))..)))).)).......-.....--- ( -36.40, z-score = -0.23, R) >droYak2.chr2L 16398189 117 + 22324452 GGAGUUUGCCACCGGCAAGACGCGACUGGUUACCGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUCUGGUUACCUAUUGCUGGUUACCUA ..((((.((((((((((.((((....(((...)))..))))))).)))))))))))((((...)))).((.(((((........))))).))..(((.(((.......))).))).. ( -41.10, z-score = -0.57, R) >droEre2.scaffold_4929 15899106 112 - 26641161 GGAGUUUGCCACCGGCAAGACGCGACUGGUUACCGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUCUGGCUAGCU-GUGCC-ACAAC--- ((((((.((((((((((.((((....(((...)))..))))))).)))))))))))((((...)))).......))..(((.((.(((.((.....)).)))-.)).)-))...--- ( -39.70, z-score = -0.13, R) >droAna3.scaffold_12916 63101 114 - 16180835 GGAGUUUGCCACCGGCAAGACGCGUCUGGUUACGGAAUGUUUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCGAAGUGUCAGGGUAAGUCU---CAAUUGAUUUAUUUUCUGGC ((((((.((((((((((((.....((((....))))...))))).)))))))))))((((...))))......)).....(((((((((((((.---.....)))))))..)))))) ( -43.30, z-score = -3.00, R) >dp4.chr4_group1 1522760 112 + 5278887 GGAGUUUGCCACCGGCAAGACGCGUCUGGUCACGGAAUGUUUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUGAGUCC--ACCAAAGAACCCCAGAAG--- .....((((((((((((((.....((((....))))...))))).)))))))))((((.((.(((.((((((......)))).))((((....)--)))..))))).))))...--- ( -41.70, z-score = -1.82, R) >droPer1.super_5 1519940 112 - 6813705 GGAGUUUGCCACCGGCAAGACGCGUCUGGUCACGGAAUGUUUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUGAGUCC--ACCAAAGAACCCCAGAAG--- .....((((((((((((((.....((((....))))...))))).)))))))))((((.((.(((.((((((......)))).))((((....)--)))..))))).))))...--- ( -41.70, z-score = -1.82, R) >droWil1.scaffold_180772 1645235 115 + 8906247 GGAGUUUGCCACCGGCAAGACGCGUCUGGUAACGGAAUGUUUGCCUGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUAAGUAA--AAAAAUAAAUCAGAUUAUAUU ..((((.((((((((((((.....((((....))))...)))))).))))))))))((((...))))(((.((((..........)))).))).--..................... ( -35.30, z-score = -2.24, R) >droVir3.scaffold_12963 3423709 109 - 20206255 GGAGUUUGCCACCGGCAAGACGCGUCUGGUUACGGAAUGUUUGCCCGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUAAGUCG------UUAGUUUCAUUCAUU-- ..((((.((((((((((((.....((((....))))...)))).)))))))))))).(((((.((.(.((.((((..........)))).)).)------..))...)))))...-- ( -35.20, z-score = -1.39, R) >droMoj3.scaffold_6500 13797588 112 - 32352404 AGAGUUUGCCACCGGCAAGACGCGUCUGGUUACGGAAUGUUUGCCUGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUAAGUCG---CGCUUAGCUCAGUUCAAU-- .((((((((((((((((((.....((((....))))...)))))).))))))))...((((((.((((((((......)))).))))......)---))))).))))........-- ( -40.30, z-score = -1.94, R) >droGri2.scaffold_15252 2987635 111 - 17193109 GGAGUUUGCCACCGGCAAGACGCGUCUGGUAACUGAAUGUUUGCCCGGUGGCAACUGGAGUGUCUCCUACAUACCAAAGUGUCAGGGUGAGUCC--GCAA--AACACAAUUCAUA-- ((((((.((((((((((((.....((........))...)))).))))))))))))((((...))))......))...((((....(((....)--))..--.))))........-- ( -34.80, z-score = -0.64, R) >anoGam1.chr2R 36295478 110 + 62725911 GGAGUUUGCGACGGGCAAGAACCGCAUCACGACCGUCUGCCAGAACGGUGGCAACUGGAGCAUCAGCUACAUCCCCGACUGUCAAGGUAAGCGA--UUGCAGGUCCGAGGUU----- (((.((((((((((.......))).........((..((((...(((((((....((.(((....))).))...)).)))))...))))..)).--))))))))))......----- ( -30.70, z-score = 1.33, R) >apiMel3.Group15 4259502 92 - 7856270 AGAAUUCGCCACCGGCAAGGCGAAAAUAACCACAGAGUGUCUUCCUAGUGGAAACUGGUCCGUCACGUACAUACCCAAUUGUCAGGAGGUGU------------------------- ....((((((........))))))....(((.....(((.(...(((((....)))))...).)))........((........)).)))..------------------------- ( -22.40, z-score = 0.02, R) >triCas2.ChLG2 10851822 92 + 12900155 AGAAUUCGCAACAGGACGUAAUAGAUUAACCACUCAGUGCCUGCCUGGUGGUAACUGGUCUGUGAGUUAUAUCCCGAAAUGUCAGGAAGUUU------------------------- .((.((((......(((...(((((((((((((.(((.(....)))))))))...))))))))..)))......))))...)).........------------------------- ( -22.80, z-score = -0.54, R) >consensus GGAGUUUGCCACCGGCAAGACGCGACUGGUUACGGAAUGUCUGCGCGGUGGCAACUGGAGUGUCUCCUACAUACCCAAGUGUCAGGGUGAGUCU___UACCUAAUUCC_AUAAU___ .....((((.....))))(((((..(..(((((..............)))))....)..))))).(((((((......)))).)))............................... (-15.39 = -15.93 + 0.53)
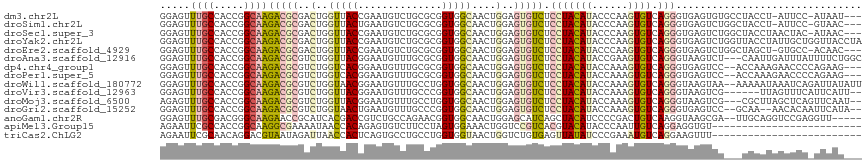
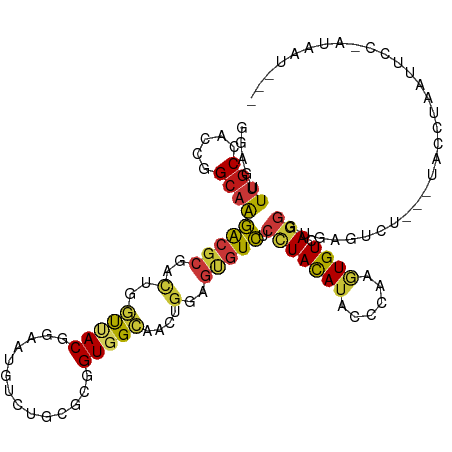
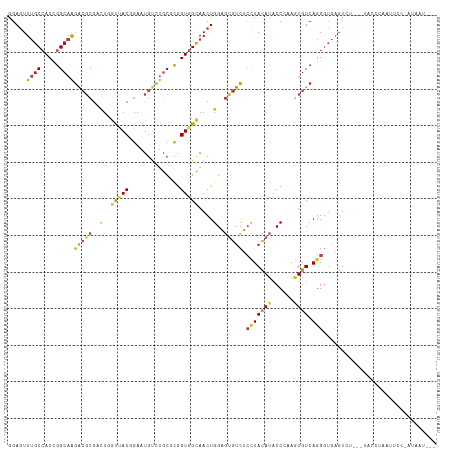
| Location | 6,983,321 – 6,983,441 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.63 |
| Shannon entropy | 0.37214 |
| G+C content | 0.53833 |
| Mean single sequence MFE | -45.14 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.59 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
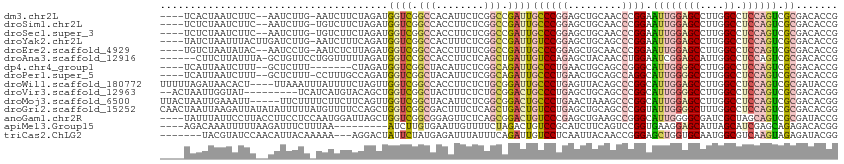
>dm3.chr2L 6983321 120 + 23011544 ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((.(((((((.(....)....((((((((((((.((((((...(((.......))).))).)))))))))).......(((((....))))).....)))))....))))))).)).. ( -44.60, z-score = -2.24, R) >droSim1.chr2L 6779209 120 + 22036055 ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCAGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((.(((((((.(....)....((((((((((((.((((((.................))).)))))))))).......(((((....))))).....)))))....))))))).)).. ( -43.33, z-score = -2.26, R) >droSec1.super_3 2516384 120 + 7220098 ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((.(((((((.(....)....((((((((((((.((((((...(((.......))).))).)))))))))).......(((((....))))).....)))))....))))))).)).. ( -44.60, z-score = -2.24, R) >droYak2.chr2L 16398226 120 - 22324452 ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((.(((((((.(....)....((((((((((((.((((((...(((.......))).))).)))))))))).......(((((....))))).....)))))....))))))).)).. ( -44.60, z-score = -2.24, R) >droEre2.scaffold_4929 15899138 120 + 26641161 ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCGGUAACCAGUCGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((.(((((((.(....)....((((((((((((.((((((...(((.......))).))).)))))))))).......(((((....))))).....)))))....))))))).)).. ( -44.60, z-score = -2.24, R) >droAna3.scaffold_12916 63135 120 + 16180835 ACUUCGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((((((((((.(....)....((((((((((((.((((((....(((......))).)).))))))))))).......(((((....))))).....)))))....)))))))))).. ( -46.50, z-score = -3.95, R) >dp4.chr4_group1 1522792 120 - 5278887 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUGACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((((((((((.(....)....((((((((((((.((((......((((......))))..))))))))))).......(((((....))))).....)))))....)))))))))).. ( -45.60, z-score = -3.36, R) >droPer1.super_5 1519972 120 + 6813705 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAAACAUUCCGUGACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG ..((((((((((.(....)....((((((((((((.((((......((((......))))..))))))))))).......(((((....))))).....)))))....)))))))))).. ( -45.60, z-score = -3.36, R) >droWil1.scaffold_180772 1645270 120 - 8906247 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCAGGCAAACAUUCCGUUACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCAAAUG ..((((((((((.(....)....(((((((((((.(((((((....((((....)))).)).))))))))))).......(((((....))))).....)))))....)))))))))).. ( -45.50, z-score = -3.56, R) >droVir3.scaffold_12963 3423738 120 + 20206255 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGGGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUGCCGAAGG .(((((((((((.(....)....(((((((((((((.(((((....(((......))).)).))))))))))).......(((((....))))).....)))))....))))))))))). ( -47.90, z-score = -3.01, R) >droMoj3.scaffold_6500 13797620 120 + 32352404 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCAGGCAAACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCUUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUGCCAAAGG .(((((((((((.(....)....(((((((((((.(((((((....(((......))).)).)))))))))))..((...(((((....))))).))..)))))....))))))))))). ( -48.70, z-score = -3.85, R) >droGri2.scaffold_15252 2987666 120 + 17193109 ACUUUGGUAUGUAGGAGACACUCCAGUUGCCACCGGGCAAACAUUCAGUUACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUGAAGCUGACCAGCGUGCCAAAGG .(((((((((((.(....)....(((((((((((((.(((((.....(((....)))..)).)))))))))))...(.(.(((((....))))).).).)))))....))))))))))). ( -48.10, z-score = -2.88, R) >anoGam1.chr2R 36295508 120 - 62725911 AGUCGGGGAUGUAGCUGAUGCUCCAGUUGCCACCGUUCUGGCAGACGGUCGUGAUGCGGUUCUUGCCCGUCGCAAACUCCUGCCCGACGGGGCAGGUAAAGUUGAUCACCGUGCCGAACG .....((...(((((((......)))))))..))((((.(((..(((((.((((((.(((....))))))))).(((((((((((....)))))))...))))....)))))))))))). ( -52.70, z-score = -2.43, R) >apiMel3.Group15 4259514 120 + 7856270 AAUUGGGUAUGUACGUGACGGACCAGUUUCCACUAGGAAGACACUCUGUGGUUAUUUUCGCCUUGCCGGUGGCGAAUUCUUGGCCAAGGGGACACGAGAACGUCACUGUUGUACCGAACG ..((.((((((.((((((((..((.((((((....)).)))).((((.((((((..((((((........))))))....)))))).))))....).)..)))))).)))))))).)).. ( -41.70, z-score = -1.65, R) >triCas2.ChLG2 10851834 120 - 12900155 AUUUCGGGAUAUAACUCACAGACCAGUUACCACCAGGCAGGCACUGAGUGGUUAAUCUAUUACGUCCUGUUGCGAAUUCUUGACCAGUCGGACAAUUAAACGUGAUGAUGGUUCCGAAAG .(((((((((.....((((.(.((.(((.......))).))).((((.(((((((...(((.(((......))))))..))))))).))))..........)))).....))))))))). ( -33.10, z-score = -1.32, R) >consensus ACUUGGGUAUGUAGGAGACACUCCAGUUGCCACCGCGCAGACAUUCCGUAACCAGACGCGUCUUGCCGGUGGCAAACUCCUGUCCAAUGGGACAUGUAAAGCUGACCAGCGUACCGAAAG .....((((((..((...........((((((((......((.....))........((.....)).)))))))).....(((((....)))))...........))..))))))..... (-25.27 = -25.59 + 0.32)
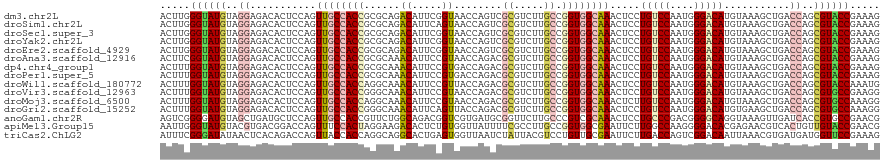
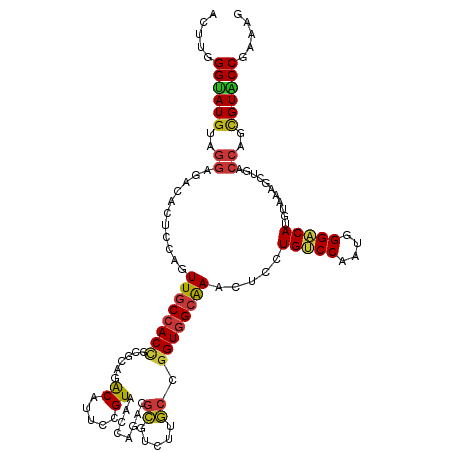
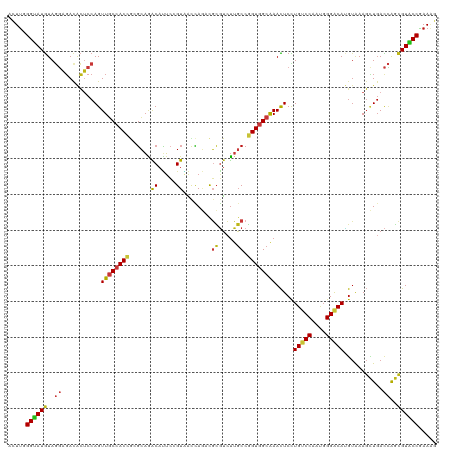
| Location | 6,983,441 – 6,983,547 |
|---|---|
| Length | 106 |
| Sequences | 15 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.01 |
| Shannon entropy | 0.77331 |
| G+C content | 0.52552 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.51 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6983441 106 - 23011544 ----UCACUAAUCUUC--AAUCUUG-AAUCUUCUAGAUGGUCGGCCACAUUCUCGGCCGAUUGCCCGGAGCUGCAACCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ----............--.((((.(-(....)).))))((((((((........))))))((((((((.(......))))).((((((((....).))))))).))))..)). ( -31.80, z-score = -1.00, R) >droSim1.chr2L 6779329 106 - 22036055 ----UCUCUAAUCUUC--AAUCUUG-UGUCUUCUAGAUGGUCGGCCACCUUCUCGGCCGAUUGCCCGGAGCUGCAACCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ----............--......(-((((........((((((((........))))))))((((((.(......))))).((((((((....).))))))).))))))).. ( -35.50, z-score = -1.74, R) >droSec1.super_3 2516504 106 - 7220098 ----UCUCUAAUCUUC--AAUCUUG-UGUCUUCUAGAUGGUCGGCCACCUUCUCGGCCGAUUGCCCGGAGCUGCAACCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ----............--......(-((((........((((((((........))))))))((((((.(......))))).((((((((....).))))))).))))))).. ( -35.50, z-score = -1.74, R) >droYak2.chr2L 16398346 108 + 22324452 ----UAUCUAAUUUACUUGAUCUUG-AAUCUUUCAGAUGGUCGGCCACUUUCUCGGCCGAUUGUCCGGAGCUGCAGCCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ----.............((.((.((-(....(((.((..(((((((........)))))))..))(((.((....))))))))(((((((....).))))))))).))))... ( -34.50, z-score = -1.29, R) >droEre2.scaffold_4929 15899258 106 - 26641161 ----UGUCUAAUAUAC--AAUCCUG-AAUCUCUUAGAUGGUCGGCCACCUUUUCGGCCGAUUGCCCGGAGCUGCAACCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ----((((........--....(((-(.....))))..((((((((........))))))))((((((.(......))))).((((((((....).))))))).))))))... ( -32.80, z-score = -1.23, R) >droAna3.scaffold_12916 63255 106 - 16180835 ------CUUCUUAUUUA-GCUGUUCCUGGUUUUUAGAUGGUCCGCCACCUUCUCAGCUGAUUGUCCAGAGCUACAACCUGGAAUCGGAGCAUUGGCCUCCAGUCGCGACACCG ------...........-(.(((..((((......((((.(((((..........)).(((..(((((.(......)))))))))))).)))).....))))..))).).... ( -23.80, z-score = 0.75, R) >dp4.chr4_group1 1522912 100 + 5278887 ----UCAUUAAUCUUU--GCUCUUU-------CUAGAUGGUCGGCUACAUUCUCGGCAGAUUGCCCUGAACUGCAGCCGGGCAUUGGGGCCUUGGCCUCCAGUCGCGACACCG ----..........((--((.....-------...((((.((((((.(((((..(((.....)))..))).)).)))))).))))(((((....))).))....))))..... ( -34.30, z-score = -1.43, R) >droPer1.super_5 1520092 106 - 6813705 ----UCAUUAAUCUUU--GCUCUUU-CCUUUGCCAGAUGGUCGGCUACAUUCUCGGCAGAUUGCCCUGAACUGCAGCCAGGCAUUGGGGCCUUGGCCUCCAGUCGCGACACCG ----......((((..--((.....-.....)).)))).(((((((.(((((..(((.....)))..))).)).))))..((((((((((....))).))))).))))).... ( -34.70, z-score = -1.10, R) >droWil1.scaffold_180772 1645390 109 + 8906247 UUUUUAGAUAACACU----UUAAAUUUAUUUUCUAGUUGGUCGGCCACCUUCUCUGCGGAUUGCCCUGAGUUACAGCCCGGCAUUGGAGCCUUGGCCUCCAGUCGCGAUACCG ......(((...(((----..(((.....)))..)))..)))(((......(((.(.((....))).))).....)))..((((((((((....).))))))).))....... ( -23.30, z-score = 0.68, R) >droVir3.scaffold_12963 3423858 102 - 20206255 --ACUAAUUGGUAU---------UCAUCAUGUACAGCUGGUCGGCUACUUUCUCUGCGGACUGCCCUGAGCUGCAGCCCGGCAUUGGGGCCUUGGCCUCCAGUCGCGACACGG --........((((---------.......))))..((((((((((.(...(((.(.((....))).)))..).))))..((((((((((....))).))))).))))).))) ( -30.80, z-score = 0.96, R) >droMoj3.scaffold_6500 13797740 108 - 32352404 UUACUAAUUGAAAUU-----UUCUUUUCUUCUUCAGUUGGUCGGCUACAUUCUCGGCGGACUGCCCUGAACUAAAGCCCGGCAUUGGAGCCUUGGCCUCCAGUCGCGACACGG ..((((((((((...-----...........)))))))))).((((...(((..(((.....)))..)))....))))..((((((((((....).))))))).))....... ( -29.94, z-score = -0.79, R) >droGri2.scaffold_15252 2987786 113 - 17193109 CAACUAAUUAAGAUUAUAUAUUUUUAUGUUUUCCAGCUGGUCGGCGACUUUCUCAGCUGACUGUCCUGAGCUGCAGCCCGGUAUUGGGGCUUUGGCCUCCAGUCGCGACACGG ...........((..(((((....)))))..))...(((((((.(((((......((..(..((((..(((((.....)))).)..)))).)..))....))))))))).))) ( -31.10, z-score = -0.34, R) >anoGam1.chr2R 36295628 109 + 62725911 ----UAUUUAUUCCUUACCUUCCUCCAAUGGAUUAGCUGGUCGGCGGAGUUCUCAGCGGACUGUCCCGAGCUGAAGCCGGGCAUUGGGGCGAUCGCUAGCAGUCGCGAUACCG ----................((((((((((........((((.((.((....)).)).))))..((((.((....)))))))))))))).))((((........))))..... ( -34.60, z-score = 0.48, R) >apiMel3.Group15 4259634 100 - 7856270 ----AGACAAAUUUUUAAGAUUUCUUUAA---------AUCUUGUGAAUUGUUUUCUAGACUGUCCGCAUCUUCAGUCCGGUGAAGGAGCAUUAGCAUCGAGCAGAGACACGG ----((((((.(((.((((((((....))---------)))))).))))))))).......(((((((..(((((......)))))..))....((.....)).).))))... ( -21.20, z-score = -0.03, R) >triCas2.ChLG2 10851954 103 + 12900155 -------UACGUAUCCAACAUUACAAAAA---AGGACUAUUCUAUGAGAUUUAUUUCAGAUUGUCCUCAAUUACAACCGGGAGCUGGUGCAAUGGCGUCAAGUAGAGAUACGG -------..((((((..............---(((((...(((..((((....)))))))..)))))...((((.(((((...)))))((....)).....)))).)))))). ( -22.80, z-score = -1.18, R) >consensus ____UAAUUAAUAUUC__GAUCUUU_UAUUUUCUAGAUGGUCGGCCACAUUCUCGGCCGAUUGCCCUGAGCUGCAGCCCGGAAUUGGAGCCUUGGCCUCCAGUCGCGACACCG ......................................((((.(((........))).))))((((((.........)))).((((((((....)).)))))).))....... (-13.28 = -13.51 + 0.23)
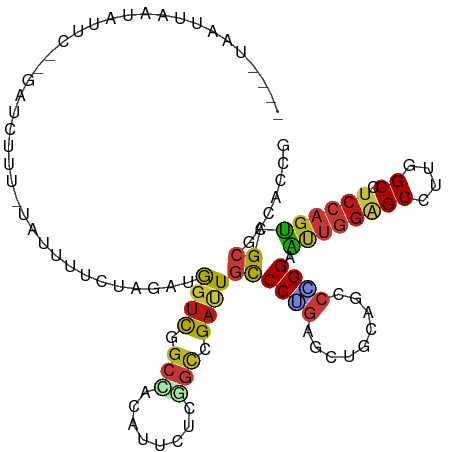
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:43 2011