
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,969,369 – 6,969,482 |
| Length | 113 |
| Max. P | 0.849729 |

| Location | 6,969,369 – 6,969,482 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.62 |
| Shannon entropy | 0.56213 |
| G+C content | 0.49730 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
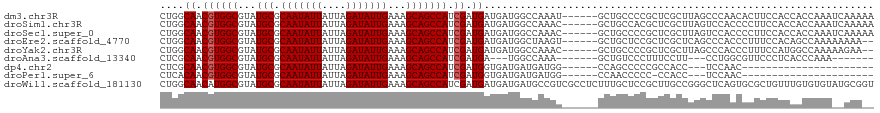
>dm3.chr3R 6969369 113 + 27905053 CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAU------GCUGCCCCGCUCGCUUAGCCCAACACUUCCACCACCAAAUCAAAAA ..(((...((((((...((.(((((((....)))))))..((((((((((......))))))....)------)))))..))).)))...))).......................... ( -28.90, z-score = -1.28, R) >droSim1.chr3R 13120296 113 - 27517382 CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAC------GCUGCCACGCUCGCUUAGUCCACCCCUUCCACCACCAAAUCAAAAA (((((..((((((...(((.(((((((....)))))))...)))((((((......)))))).....------...))))))...)).)))............................ ( -30.90, z-score = -2.09, R) >droSec1.super_0 6131819 113 - 21120651 CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAC------GCUGCCCCGCUCGCUUAGUCCACCCCUUCCACCACCAAAUCAAAAA ..(((...(..((((.(((.(((((((....)))))))...)))((((((......))))))...))------))..)...)))................................... ( -28.90, z-score = -1.58, R) >droEre2.scaffold_4770 3099443 111 - 17746568 CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCUAAGU------GCUGCUCCGCUCGCUCAGCCCACCCUUUCCACAGCCAAAAAAAA-- .((((...((((.(((.((((((((((....))))))......(((((((......)))))))..))------))))).))))..((...))..............)))).......-- ( -32.10, z-score = -1.46, R) >droYak2.chr3R 11001042 111 + 28832112 CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAC------GCUGCCCCGCUCGCUUAGCCCACCCUUUCCAUGGCCAAAAAGAA-- .((((...(..((((.(((.(((((((....)))))))...)))((((((......))))))...))------))..)...(((.....)))..............)))).......-- ( -34.00, z-score = -1.85, R) >droAna3.scaffold_13340 11530658 99 - 23697760 CUCGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGA---UGGCCAAA-------GCUGUCCCUUUCCUU---CCUGGCGUUCCCUCACCCAAA------- ..(((..((((((...(((.(((((((....)))))))...))))))).))..((---((((....-------)))))).........---....)))..............------- ( -24.00, z-score = -0.85, R) >dp4.chr2 28474057 88 + 30794189 CUCGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGGUGAUGAUGAUGG------CCAGCCCCCGCCACC---UCCAAC---------------------- ........((((((..(((.(((((((....)))))))...)))(((((((.(......).))))))------).......)))))).---......---------------------- ( -27.40, z-score = -1.99, R) >droPer1.super_6 3804482 87 + 6141320 CUCACAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGGUGAUGAUGAUGG------CCAACCCCC-CCACC---UCCAAC---------------------- ........((((....(((.(((((((....)))))))...)))(((((((.(......).))))))------)........-)))).---......---------------------- ( -21.40, z-score = -1.25, R) >droWil1.scaffold_181130 7704751 119 + 16660200 CUGGCAACAUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGAUGCCGUCGCCUCUUUGCUCCGCUUGCCGGGCUCAGUGCGCUGUUUGUGUGUAUGCGGU ..(((...(((((((.(((.(((((((....)))))))...)))..((((......))))))))))).)))........((((.(((((.(..(((....)))..).)).))).)))). ( -37.30, z-score = 0.56, R) >consensus CUGGCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAC______GCUGCCCCGCUCGCUUAGCCCACCCCUUCCACCACCAAA__AAA__ ....((.((((((...(((.(((((((....)))))))...))))))).)).))................................................................. (-15.36 = -15.47 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:21 2011