
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,918,536 – 6,918,676 |
| Length | 140 |
| Max. P | 0.620610 |

| Location | 6,918,536 – 6,918,638 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Shannon entropy | 0.36575 |
| G+C content | 0.44798 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -14.28 |
| Energy contribution | -16.33 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
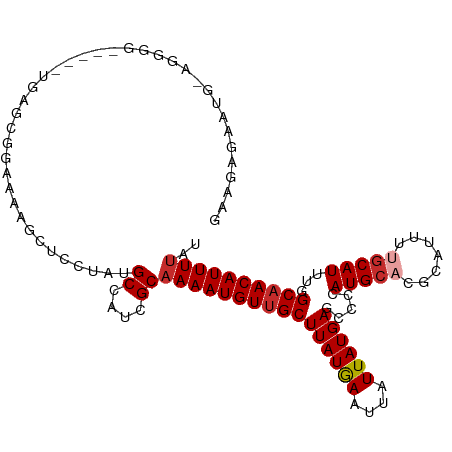
>dm3.chr3R 6918536 102 - 27905053 CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGGAGCUUUUCCGCUCACCCCUCCAUUCUGUUCACAUGCCACUACGCGAAUUUGCUUGCGUGUGCUU-------- ..........((((.((((((.((((((.((((((..((((......)))).............(....)))))))...))))))..)).)))).))))...-------- ( -25.60, z-score = -1.85, R) >droSim1.chr3R 13067887 99 + 27517382 CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGGAGCUUUUCCGCUCACCCCUGCAUUCUGUUCACUUGCCACUACGCGAAUUUGCUUGCGUGUG----------- ..........((((.((((((.((((((.(((((...((((......((........)).....))))...)))))...))))))..)).)))).))))----------- ( -26.60, z-score = -2.20, R) >droSec1.super_0 6075195 102 + 21120651 CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGGAGCUUUUCCGCUCACCCCUCCAUUCUGUUCACUUGCCACUACGCGAAUUUGCUUGCGUGUGCUU-------- ..........((((.((((((.((((((.(((((.((((((......))))....(........)....)))))))...))))))..)).)))).))))...-------- ( -24.30, z-score = -1.53, R) >droYak2.chr3R 10949076 109 - 28832112 CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGUAGCUUUUCCGCUCACCCCU-CAUUCUCUACACUUGCCACUACGCGAAUUUGCUAGCGUGUGUACAUGUGUGU ..............(((((...((((((.(((((.((((((......))).......-..........))))))))...)))))).))))).(((((....))))).... ( -23.12, z-score = -0.60, R) >droEre2.scaffold_4770 3048669 96 + 17746568 CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGGAGCAUUUCCGCUCACCCU--CAUUCUCUUCACUUGCCACUACGCGAAUUUGCUUGUGUAU------------ ..........(((((((((...((((((.(((((...((((......)))).....--.............)))))...)))))).)))))))))...------------ ( -26.90, z-score = -3.97, R) >droAna3.scaffold_13340 11479265 91 + 23697760 CAUAAUAAUUCAUAAGCAACAUUUUGCCAUGGCA---GAGCGGAGCAGCCUGCCUC----UACUGGUACUUCUCCUUCACCCCUAUUCCCUGGUGCAU------------ ...............(((.((((((((....)))---))).((((.((..((((..----....)))))).))))...............)).)))..------------ ( -20.00, z-score = -0.21, R) >consensus CAUAAUAAUUCAUAAGCAACAUUUUGCGAUGGCAUAGGAGCUUUUCCGCUCACCCCU_CAUUCUGUUCACUUGCCACUACGCGAAUUUGCUUGCGUGUG___________ ............(((((((...((((((.(((((...((((......))))....................)))))...)))))).)))))))................. (-14.28 = -16.33 + 2.06)


| Location | 6,918,570 – 6,918,676 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Shannon entropy | 0.51957 |
| G+C content | 0.44362 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.09 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6918570 106 + 27905053 GAACAGAAUGGAGGGG-----UGAGCGGAAAAGCUCCUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ....((((((....((-----.((((......)))))).(((((..(((((((((.((((((((....)))))))).........)))))))))...))))).)))))).. ( -33.40, z-score = -2.31, R) >droSim1.chr3R 13067918 106 - 27517382 GAACAGAAUGCAGGGG-----UGAGCGGAAAAGCUCCUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ....((((((....((-----.((((......)))))).(((((..(((((((((.((((((((....)))))))).........)))))))))...))))).)))))).. ( -32.50, z-score = -1.88, R) >droSec1.super_0 6075229 106 - 21120651 GAACAGAAUGGAGGGG-----UGAGCGGAAAAGCUCCUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ....((((((....((-----.((((......)))))).(((((..(((((((((.((((((((....)))))))).........)))))))))...))))).)))))).. ( -33.40, z-score = -2.31, R) >droYak2.chr3R 10949118 105 + 28832112 GUAGAGAAUG-AGGGG-----UGAGCGGAAAAGCUACUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ....((((((-...((-----(.(((......)))))).(((((..(((((((((.((((((((....)))))))).........)))))))))...))))).)))))).. ( -31.10, z-score = -2.03, R) >droEre2.scaffold_4770 3048699 104 - 17746568 GAAGAGAAUG--AGGG-----UGAGCGGAAAUGCUCCUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ....((((((--..((-----.(((((....))))))).(((((..(((((((((.((((((((....)))))))).........)))))))))...))))).)))))).. ( -33.20, z-score = -2.72, R) >droAna3.scaffold_13340 11479289 105 - 23697760 GAGAAGUACC-AGUAG-----AGGCAGGCUGCUCCGCUCUGCCAUGGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCUACAUUUUAU ((((.(((((-((...-----.(((((((......)).)))))...(((((((((.((((((((....)))))))).........)))))))))..)))).))).)))).. ( -32.90, z-score = -1.68, R) >dp4.chr2 28402094 98 + 30794189 GAGGGGUGCGGGGGGG--------CCAUUGGGGGGGUUCUGCCAUUGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCC-AUGC-CGCAC---GCAUUAUGCAACAUUUUAC ...(.((((((.((((--------(((((((.((....)).)))..((((....))))............))).))))-.).)-)))))---.)................. ( -30.70, z-score = -0.10, R) >droPer1.super_6 3738917 106 + 6141320 GAGGGGUGCGGGGGGGGGGUGCGGCCCCAGCAAAUGUUCUGCCAUUGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCC-AUGC-CGCAC---GCAUUAUGCAACAUUUUAC ...(.((((((.(((((((.....))))....((((((.(((....))).))))))((((((((....))))))))))-.).)-)))))---.)................. ( -34.70, z-score = 0.28, R) >droWil1.scaffold_181130 7654010 93 + 16660200 ----------------CACCAACACUGCAGGAUGCACGCUGCCAUUGCAAAAUGUUGCUUAUAAAUUAUUAUGAGCC--AUGGCCAUGCCACGCAUUUUGCAACAUUUAAU ----------------..........((((........))))..(((((((((((.((((((((....)))))))).--.((((...)))).)))))))))))........ ( -26.70, z-score = -1.87, R) >consensus GAAGAGAAUG_AGGGG_____UGAGCGGAAAAGCUCCUAUGCCAUCGCAAAAUGUUGCUUAUGAAUUAUUAUGAGCCCCAUGCACGCAUUUUGCAUUUGGCAACAUUUUAU ........................................((....))((((((((((((((((....)))))).....(((((.......)))))...)))))))))).. (-13.69 = -14.09 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:16 2011