
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,915,033 – 6,915,149 |
| Length | 116 |
| Max. P | 0.998999 |

| Location | 6,915,033 – 6,915,149 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.44687 |
| G+C content | 0.28138 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.998999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
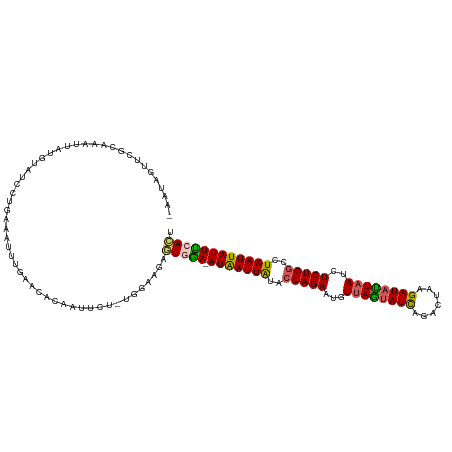
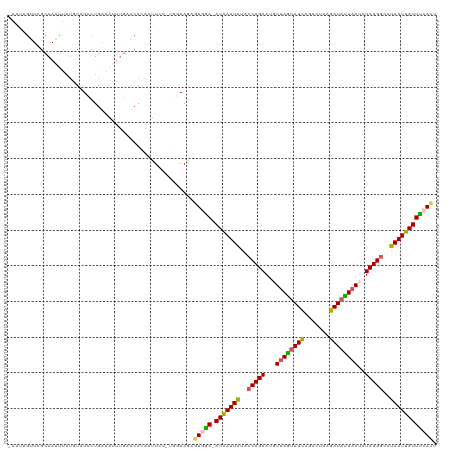
>dm3.chr3R 6915033 116 + 27905053 UAUUGUUUUGCAAGCUAUGUAUCCUGAAAUUUGAACACAAUU---UGGAAGUGUGAA-AUAAUUAAACCAGAAUGUUAGUAUCAGACUAAGAUAUUAAUCUCUGGCCUAAUUAUUUCACU ........((((.....))))(((......(((....)))..---.)))...(((((-(((((((..(((((..(((((((((.......))))))))).)))))..)))))))))))). ( -29.00, z-score = -3.53, R) >droSim1.chr3R 13063630 119 - 27517382 AAGUUGUUGGCAAAUUAUAUAUCCUGAAAUUUGAACAGAAUUGUUUGGAAGUGUGGA-AUAAUUAUACCAGAAUGUUAGUAUCAGACUAAGAUAUUAAUCUCUGGUCUAAUUAUUUCACU ...(((....)))................((..(((......)))..))...(((((-(((((((.((((((..(((((((((.......))))))))).)))))).)))))))))))). ( -32.90, z-score = -3.94, R) >droSec1.super_0 6071837 118 - 21120651 AAAUUGUUGGCAAAUUAAAUAUCCUGAAAUUUGAACAGAAUUGUUUGGAAGAGUGGAUAUAAUUA--CCAGAAUGUUAGUAUCAGAAUAAGAUAUUAAUCUCUGUCCUAAUUAUUUCACU ((((.(((((........((((((.....((..(((......)))..)).....)))))).....--.((((..(((((((((.......))))))))).))))..))))).)))).... ( -23.00, z-score = -1.29, R) >droYak2.chr3R 10945535 105 + 28832112 -UAAAGUUCGCAAAUAAUGUAUUCUG---------CACAUUUCU----AAAAAUAGA-AUGAUUGUACCAGAAGUUUAUUAUCAGACUAAGAUAAUAACUUCUGGCCUAAUUAUUCCAAU -........(((((((...)))).))---------)........----.......((-(((((((..(((((((((.((((((.......)))))))))))))))..))))))))).... ( -26.70, z-score = -5.04, R) >droEre2.scaffold_4770 3045230 114 - 17746568 -UAAAGUUCGCAAAUAUUGUAUUCUGAAAUUUGGGCACAUUUCU----AAGAAUAGA-AUGAUUAUUCCAGAAGUUUAGUAUUAGACUAUGAUUCUUAUUUCUGGCCUAAUUAUUCCAUA -..................((((((......((....)).....----.))))))((-(((((((..((((((((..((.((((.....)))).)).))))))))..))))))))).... ( -24.30, z-score = -1.89, R) >consensus _AAUAGUUCGCAAAUUAUGUAUCCUGAAAUUUGAACACAAUUCU_UGGAAGAGUGGA_AUAAUUAUACCAGAAUGUUAGUAUCAGACUAAGAUAUUAAUCUCUGGCCUAAUUAUUUCACU ....................................................(((((.(((((((..(((((...((((((((.......))))))))..)))))..)))))))))))). (-17.54 = -17.10 + -0.44)

| Location | 6,915,033 – 6,915,149 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.44687 |
| G+C content | 0.28138 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.52 |
| Energy contribution | -16.56 |
| Covariance contribution | 3.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
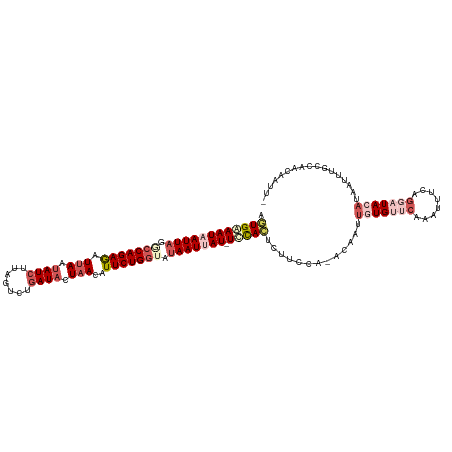
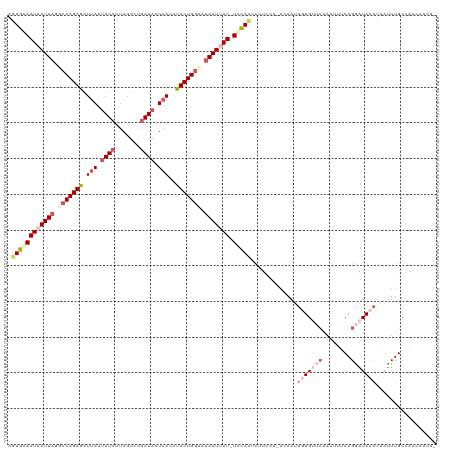
>dm3.chr3R 6915033 116 - 27905053 AGUGAAAUAAUUAGGCCAGAGAUUAAUAUCUUAGUCUGAUACUAACAUUCUGGUUUAAUUAU-UUCACACUUCCA---AAUUGUGUUCAAAUUUCAGGAUACAUAGCUUGCAAAACAAUA .((((((((((((((((((((.(((.((((.......)))).)))..)))))))))))))))-))))).......---...(((((((........)))))))................. ( -34.60, z-score = -5.87, R) >droSim1.chr3R 13063630 119 + 27517382 AGUGAAAUAAUUAGACCAGAGAUUAAUAUCUUAGUCUGAUACUAACAUUCUGGUAUAAUUAU-UCCACACUUCCAAACAAUUCUGUUCAAAUUUCAGGAUAUAUAAUUUGCCAACAACUU .(((.((((((((.(((((((.(((.((((.......)))).)))..))))))).)))))))-).)))...........((((((.........)))))).................... ( -25.10, z-score = -3.45, R) >droSec1.super_0 6071837 118 + 21120651 AGUGAAAUAAUUAGGACAGAGAUUAAUAUCUUAUUCUGAUACUAACAUUCUGG--UAAUUAUAUCCACUCUUCCAAACAAUUCUGUUCAAAUUUCAGGAUAUUUAAUUUGCCAACAAUUU .........(((((((..(((((....))))).)))))))..........(((--(((..((((((.........((((....)))).........)))))).....))))))....... ( -19.27, z-score = -1.18, R) >droYak2.chr3R 10945535 105 - 28832112 AUUGGAAUAAUUAGGCCAGAAGUUAUUAUCUUAGUCUGAUAAUAAACUUCUGGUACAAUCAU-UCUAUUUUU----AGAAAUGUG---------CAGAAUACAUUAUUUGCGAACUUUA- ..((((((.(((..((((((((((((((((.......)))))).))))))))))..))).))-)))).....----.......((---------((((........)))))).......- ( -29.30, z-score = -4.59, R) >droEre2.scaffold_4770 3045230 114 + 17746568 UAUGGAAUAAUUAGGCCAGAAAUAAGAAUCAUAGUCUAAUACUAAACUUCUGGAAUAAUCAU-UCUAUUCUU----AGAAAUGUGCCCAAAUUUCAGAAUACAAUAUUUGCGAACUUUA- ...(((((.((((..((((((..........................))))))..)))).))-)))(((((.----.(((((........))))))))))...................- ( -17.47, z-score = -0.54, R) >consensus AGUGAAAUAAUUAGGCCAGAGAUUAAUAUCUUAGUCUGAUACUAACAUUCUGGUAUAAUUAU_UCCACUCUUCCA_ACAAUUGUGUUCAAAUUUCAGGAUACAUAAUUUGCCAACAAUU_ .((((((((((((.(((((((.(((.((((.......)))).)))..))))))).))))))).))))).............(((((((........)))))))................. (-13.52 = -16.56 + 3.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:14 2011