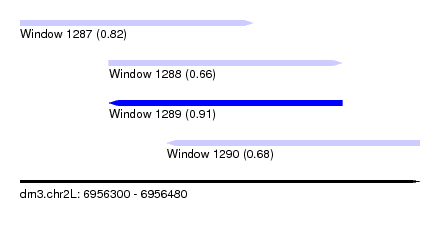
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,956,300 – 6,956,480 |
| Length | 180 |
| Max. P | 0.911529 |

| Location | 6,956,300 – 6,956,405 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Shannon entropy | 0.19875 |
| G+C content | 0.37218 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -10.40 |
| Energy contribution | -12.73 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6956300 105 + 23011544 ACAGUACAUUUUUAAAACCUCCUUAACUACACUUUAUAAACACAACACAUGUUUGCCGAUCUAGCCAAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAUUA (((((.............................................(((((.(......).)))))((((((....))))))....))))).......... ( -18.10, z-score = -2.75, R) >droSim1.chr2L 6753211 102 + 22036055 AUAGUACAUUUACAAAACCUCCUUAACACAACACAAUAAACCUAUGUCAAUUGUG---AUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAUUA ((((((.........................((((((..((....))..))))))---...((((.....((((((....))))))....)))).)))))).... ( -20.20, z-score = -3.56, R) >droSec1.super_3 2488995 102 + 7220098 AUAGUACAUUUUCAAAACCUCCUUAACACAACACAAUAAACCUAUGCCAAUUGUG---AUCUAGCCCAACACGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAUUA ((((((.........................((((((............))))))---...((((......(((((....))))).....)))).)))))).... ( -15.30, z-score = -2.15, R) >consensus AUAGUACAUUUUCAAAACCUCCUUAACACAACACAAUAAACCUAUGACAAUUGUG___AUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAUUA ((((((.........................((((((............))))))......((((.....((((((....))))))....)))).)))))).... (-10.40 = -12.73 + 2.33)

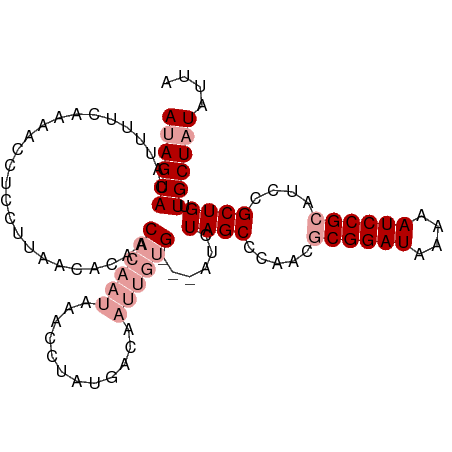
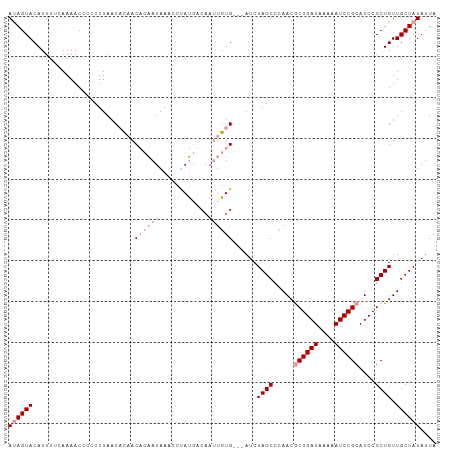
| Location | 6,956,340 – 6,956,445 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.16 |
| Shannon entropy | 0.51940 |
| G+C content | 0.33826 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.42 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6956340 105 + 23011544 ------CACAACACAU--GUUUGCC------GAUCUAGCCAAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAU-UAGUAAAAAGUUUUAAAUAAGAGAAGGCAUAAAUUACUUUAU ------..........--...((((------..((((((.....((((((....))))))....))).((((((...-)))))).............)))...))))............. ( -21.30, z-score = -1.81, R) >droSim1.chr2L 6753242 111 + 22036055 ------CACAAUAAAC--CUAUGUCAAUUGUGAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAU-UAGUAAAAAGUAUUAAAUAAGAGAAGGAUUAAAUUACUUUUG ------..........--......(((..(((((.((..((...((((((....))))))....(((.((((((...-))))))..)))..............))..)).)))))..))) ( -21.40, z-score = -2.07, R) >droSec1.super_3 2489026 111 + 7220098 ------CACAAUAAAC--CUAUGCCAAUUGUGAUCUAGCCCAACACGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAU-UAGUAAAAAGUAUUAAAUAGGAGAAUGAUUAAAUUACUAUUG ------..........--......((((.(((((.(((..((((((((((..........))))).)))))((((.(-(((((.....))))))))))........))).))))).)))) ( -17.50, z-score = -0.96, R) >droYak2.chr2L 16371265 119 - 22324452 CGUUAAUAAGGUU-UUUGGUUAAACACCUACAUUUACGACCCAUGCGGAUAAAAAUCCGCAUCCGCUGUAUCUAUAUAUAGUGAUAAGUAUUAGAUAAGAAAAUGUACAAAUCACUUUUG .........((((-(..(((.....)))(((((((.(.....((((((((....)))))))).((((((((....))))))))...............).))))))).)))))....... ( -25.10, z-score = -2.21, R) >droEre2.scaffold_4929 15872346 119 + 26641161 AGGUAUUUGGGUAGUUUGGUUAAUCAUCUACUUUCUUGACCCAUGCGAAUAAAAAUCCGCAUCCGCUGUAGCUAU-UUUAGUUAUAGGUAUUAAAUAAGAAGAUGUUUCGAUCACUUUUG ................(((((...(((((.(((..((((..((((((..........)))))...(((((((((.-..))))))))))..))))..))).)))))....)))))...... ( -22.80, z-score = -0.28, R) >consensus ______CACAAUAAAU__GUUUGCCAACUACGAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUAU_UAGUAAAAAGUAUUAAAUAAGAGAAUGAUUAAAUUACUUUUG .................................(((........((((((....))))))....(((..(((((....)))))...)))........))).................... (-10.10 = -10.42 + 0.32)

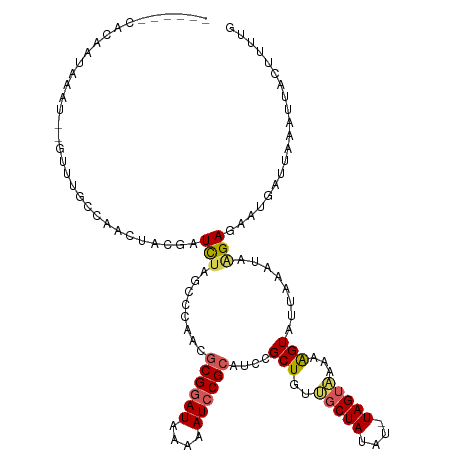
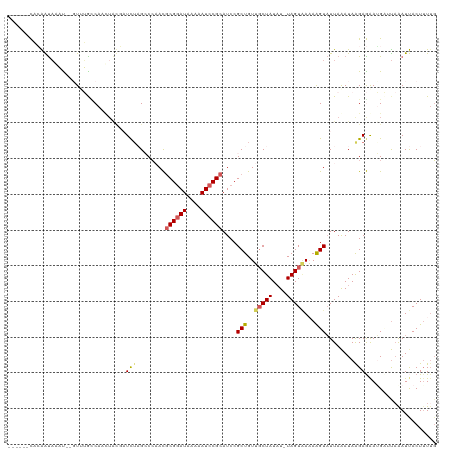
| Location | 6,956,340 – 6,956,445 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.16 |
| Shannon entropy | 0.51940 |
| G+C content | 0.33826 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -11.81 |
| Energy contribution | -11.61 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6956340 105 - 23011544 AUAAAGUAAUUUAUGCCUUCUCUUAUUUAAAACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUUGGCUAGAUC------GGCAAAC--AUGUGUUGUG------ (((((....)))))............................-.....((((((.(((((((((((....)))))))))))(((.....------)))....--...)))))).------ ( -24.80, z-score = -1.87, R) >droSim1.chr2L 6753242 111 - 22036055 CAAAAGUAAUUUAAUCCUUCUCUUAUUUAAUACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUGGGCUAGAUCACAAUUGACAUAG--GUUUAUUGUG------ ........(((((..((.........................-.....((....)).(((((((((....)))))))))))..)))))((((((.(((....--))).))))))------ ( -24.70, z-score = -2.43, R) >droSec1.super_3 2489026 111 - 7220098 CAAUAGUAAUUUAAUCAUUCUCCUAUUUAAUACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGUGUUGGGCUAGAUCACAAUUGGCAUAG--GUUUAUUGUG------ ...((((((...........................))))))-...((((..((((....((((((....)))))))))).))))...((((((..((....--))..))))))------ ( -21.53, z-score = -0.89, R) >droYak2.chr2L 16371265 119 + 22324452 CAAAAGUGAUUUGUACAUUUUCUUAUCUAAUACUUAUCACUAUAUAUAGAUACAGCGGAUGCGGAUUUUUAUCCGCAUGGGUCGUAAAUGUAGGUGUUUAACCAAA-AACCUUAUUAACG ....((((((..(((...............)))..))))))...((((..(((.((..((((((((....))))))))..)).)))..))))(((.....)))...-............. ( -27.56, z-score = -2.53, R) >droEre2.scaffold_4929 15872346 119 - 26641161 CAAAAGUGAUCGAAACAUCUUCUUAUUUAAUACCUAUAACUAAA-AUAGCUACAGCGGAUGCGGAUUUUUAUUCGCAUGGGUCAAGAAAGUAGAUGAUUAACCAAACUACCCAAAUACCU ....(((.....((.(((((.(((.(((...((((((.......-...((....))....((((((....)))))))))))).))).))).))))).))......)))............ ( -21.20, z-score = -1.35, R) >consensus CAAAAGUAAUUUAAACAUUCUCUUAUUUAAUACUUUUUACUA_AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUGGGCUAGAUCACAAUUGACAAAC__AUUUAUUGUG______ ....(((((...........................)))))..............(.(((((((((....))))))))).)....................................... (-11.81 = -11.61 + -0.20)

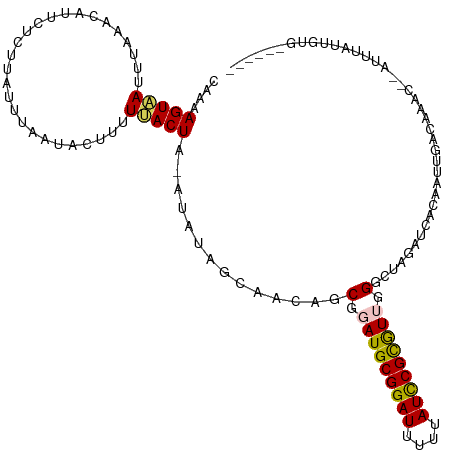
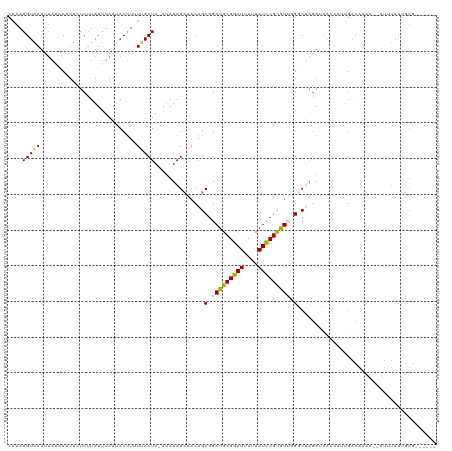
| Location | 6,956,366 – 6,956,480 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Shannon entropy | 0.43542 |
| G+C content | 0.30660 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -11.69 |
| Energy contribution | -11.49 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6956366 114 - 23011544 AAGUUUCCAUUCAAAUAUAGUUUCUUUGGUAGUUCAUAAAGUAAUUUAUGCCUUCUCUUAUUUAAAACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUU ...................(((...(((((((..((((((....)))))).....................))))))-)...)))......((((((((((....)))))))))) ( -20.20, z-score = -1.53, R) >droSim1.chr2L 6753274 103 - 22036055 -------AACUUAACUUUCCAUUCUUU----GUUCCAAAAGUAAUUUAAUCCUUCUCUUAUUUAAUACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUG -------....((((..(((.......----((...(((((((..((((............))))))))))).))..-.....((....))))).((((((....)))))))))) ( -16.80, z-score = -2.54, R) >droSec1.super_3 2489058 102 - 7220098 ------------AAGUUUUCAUUCUUUGGUAGUUCCAAUAGUAAUUUAAUCAUUCUCCUAUUUAAUACUUUUUACUA-AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGUGUUG ------------.............((((.....))))((((((...........................))))))-.....((....)).(((((((((....))))))))). ( -15.53, z-score = -0.93, R) >droYak2.chr2L 16371304 115 + 22324452 AAAUUUAAAUUGAAAUAUAAUUUCAGUGGGCGUUCCAAAAGUGAUUUGUACAUUUUCUUAUCUAAUACUUAUCACUAUAUAUAGAUACAGCGGAUGCGGAUUUUUAUCCGCAUGG ..(((((.((((((((...))))))))((.....))...((((((..(((...............)))..)))))).....))))).......((((((((....)))))))).. ( -23.06, z-score = -1.52, R) >droEre2.scaffold_4929 15872386 114 - 26641161 AAAUUUCAAUUCAAAUAUAAUUUCAGUGGGAGUUCCAAAAGUGAUCGAAACAUCUUCUUAUUUAAUACCUAUAACUAAA-AUAGCUACAGCGGAUGCGGAUUUUUAUUCGCAUGG .........................(((((.(((....(((.(((......)))..)))....))).))))).......-...((....))..((((((((....)))))))).. ( -15.90, z-score = 0.29, R) >consensus AA_UUU_AAUUCAAAUAUAAUUUCUUUGGGAGUUCCAAAAGUAAUUUAAACAUUCUCUUAUUUAAUACUUUUUACUA_AUAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUG .................................(((...(((((...........................))))).......((....)))))(((((((....)))))))... (-11.69 = -11.49 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:40 2011