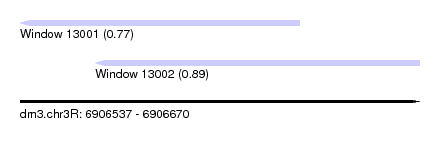
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,906,537 – 6,906,670 |
| Length | 133 |
| Max. P | 0.889995 |

| Location | 6,906,537 – 6,906,630 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Shannon entropy | 0.63559 |
| G+C content | 0.43577 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -7.70 |
| Energy contribution | -7.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6906537 93 - 27905053 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU-----UCGGUUUGG--CCAA-CCGGCUUCA--CUCGCCGCCUCAUCCUUUAUAUCCUUC- .....(((((((........))))))).......((((..((((-----..(((..((--((..-..))))..)--)).))))..))))..............- ( -23.50, z-score = -2.64, R) >droEre2.scaffold_4770 3036065 80 + 17746568 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU-----UCGGUUUGG--CCAA-CCG---GCU--CAUCCU-UUAUAUCCUUC---------- .....(((((((........))))))).......((((..((((-----(.((.....--))))-)))---..)--)))...-...........---------- ( -21.20, z-score = -2.92, R) >droYak2.chr3R 10936710 85 - 28832112 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUACGGUUCGGUUUGG--CCGA-CCG---GCU--CAUCCU-UUAUAUCCUUC---------- .....(((((((........))))))).......((((..((((.(((((......))--))))-)))---..)--)))...-...........---------- ( -24.00, z-score = -2.49, R) >droSec1.super_0 6063306 93 + 21120651 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU-----UCGGUUUGG--CCGG-CCGGCUUCA--CUCGCCGCCUCAUCCUUUAUAUCCUUC- .....(((((((........))))))).......((((..((((-----..(((..((--((..-..))))..)--)).))))..))))..............- ( -24.00, z-score = -1.73, R) >droSim1.chr3R 13056170 93 + 27517382 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU-----UCGGUUUGG--CCGG-CCGGCUUCA--CUCGCCGCCUCAUCCUUUAUAUCCUUC- .....(((((((........))))))).......((((..((((-----..(((..((--((..-..))))..)--)).))))..))))..............- ( -24.00, z-score = -1.73, R) >droAna3.scaffold_13340 11467016 96 + 23697760 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU-----UUGGUUUGG--CUGG-ACUCCCUCAUCCUUCCCACUCUUUUCCCUUAACUCAUUC .....(((((((........)))))))....((((......((.-----..((..(((--..((-(........)))...)))..))...)).......)))). ( -16.32, z-score = -1.07, R) >dp4.chr2 28384888 94 - 30794189 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUUUUACCCGGUUUGG--UCGG-CCAGCCGCU--CAUCCUAUCAUAUCCAUCCAACC----- .....(((((((........))))))).....(((((((((((......))))).((.--.(((-....)))..--)).....))))))..........----- ( -21.10, z-score = -1.32, R) >droPer1.super_6 3722540 94 - 6141320 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUUUUACCCGGUUUGG--UCGG-CCAGCCGCU--CAUCCUAUCAUAUCCAUCCAACC----- .....(((((((........))))))).....(((((((((((......))))).((.--.(((-....)))..--)).....))))))..........----- ( -21.10, z-score = -1.32, R) >droWil1.scaffold_181130 7639769 103 - 16660200 UUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUUGGACUGUCUCCGGAUUCGGUUCAAUCUCUCCAUCCUUAUCCUCAUCUUCAUGUCCAUU- ......((((((.((((((((.((....)).))))))))..((((((((((((....))..)))))))))).....................)))))).....- ( -22.00, z-score = -1.88, R) >droVir3.scaffold_13047 14231756 81 + 19223366 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGCGAAUCG-------CUUGCCCAG--CCAAUGUCGUCGCU--CAUUUUGAUGUUGUUG------------ ..((((((((((........))))))(((((((.((((...(-------((((.....--.))).))..)))).--....))))))).))))------------ ( -17.50, z-score = -0.44, R) >droMoj3.scaffold_6540 21434999 81 - 34148556 CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUACUGCGAAUCG-------CUUGCCCAG--CCAAUGUCGUCUCU--CAUUUUGUUGUUGUUG------------ ..((((((((((........))))))(((.....(((...))-------).))).(((--(.((((........--))))..))))..))))------------ ( -13.40, z-score = 0.04, R) >consensus CUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGU_____UCGGUUUGG__CCGG_CCGGCCUCU__CAUCCUGUCAUAUCCUUC__A_C_____ .....(((((((........)))))))............................................................................. ( -7.70 = -7.70 + 0.00)

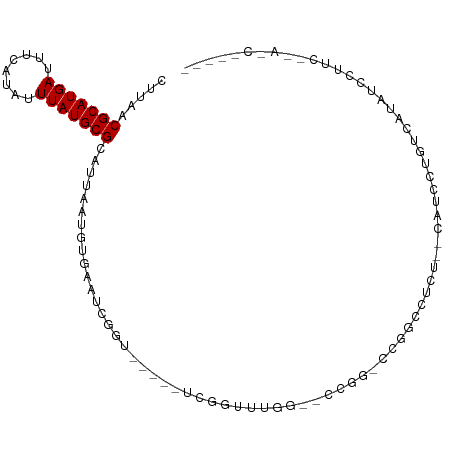
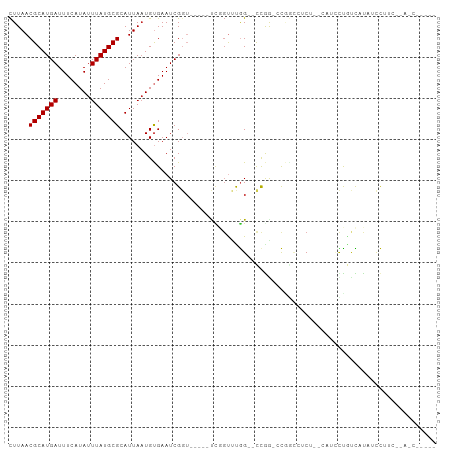
| Location | 6,906,562 – 6,906,670 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Shannon entropy | 0.35261 |
| G+C content | 0.35840 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6906562 108 - 27905053 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAU-----CGGUUCGGUUUGGCCAACCGGCUUCACU-------- ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))((((((-----(((((.((.....)))))))).))))).-------- ( -32.70, z-score = -3.72, R) >droEre2.scaffold_4770 3036082 103 + 17746568 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAU-----CGGUUCGGUUUGGCCAACCGGCU------------- (((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))..(-----(((((.((.....))))))))..------------- ( -30.50, z-score = -3.18, R) >droYak2.chr3R 10936727 108 - 28832112 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUACGGUUCGGUUUGGCCGACCGGCU------------- (((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))..(((((.(((((......))))))))))..------------- ( -33.30, z-score = -3.15, R) >droSec1.super_0 6063331 108 + 21120651 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAU-----CGGUUCGGUUUGGCCGGCCGGCUUCACU-------- ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))((((((-----((((.(((.....)))))))).))))).-------- ( -33.90, z-score = -3.54, R) >droSim1.chr3R 13056195 108 + 27517382 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAU-----CGGUUCGGUUUGGCCGGCCGGCUUCACU-------- ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))((((((-----((((.(((.....)))))))).))))).-------- ( -33.90, z-score = -3.54, R) >droAna3.scaffold_13340 11467044 108 + 23697760 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAU-----CGGUUUGGUUUGGCUGGACUCCCUCAUC-------- (((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))...-----.((((..(.....)..))))........-------- ( -27.30, z-score = -2.28, R) >dp4.chr2 28384913 109 - 30794189 CAUGUUAUUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUUUUACCCGGUUUGGUCGGCCAGCCG------------ (((((((.(((..(((((..(((((.........))))).))))).((((((........))))))))).)))))))....((.....))(((((.((....)))))))------------ ( -26.50, z-score = -1.31, R) >droPer1.super_6 3722565 109 - 6141320 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUUUUACCCGGUUUGGUCGGCCAGCCG------------ (((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))....((.....))(((((.((....)))))))------------ ( -29.70, z-score = -2.31, R) >droWil1.scaffold_181130 7639793 119 - 16660200 CAUGGUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUUUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG--GUUGGACUGUCUCCGGAUUCGGUUCAAUCUCUCCAUC .((((((((((...............((((((........))))))((((((........)))))))))))).........(--(((((((((((....))..))))))))))...)))). ( -30.30, z-score = -2.21, R) >droVir3.scaffold_13047 14231775 102 + 19223366 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGCGAAUCG--CUUGCCCAGCCAAUGUCGUC----------------- ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))((((...(--((.....))).....)))).----------------- ( -22.00, z-score = -0.80, R) >droMoj3.scaffold_6540 21435018 102 - 34148556 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUACUGCGAAUCG--CUUGCCCAGCCAAUGUCGUC----------------- .....((((((..(((((..(((((.........))))).))))).((((((........))))))))))))..((((...(--((.....))).....)))).----------------- ( -19.50, z-score = 0.03, R) >droGri2.scaffold_14906 3772852 102 + 14172833 CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGCGAAUCG--CUUG-CCAGCCAAUGUCGUCA---------------- ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))((((...(--((..-..))).....))))..---------------- ( -22.00, z-score = -0.39, R) >consensus CAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG___CGGUUCGGUUUGGCCGGCCGGCU_____________ ...((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))............................................... (-16.89 = -17.07 + 0.17)
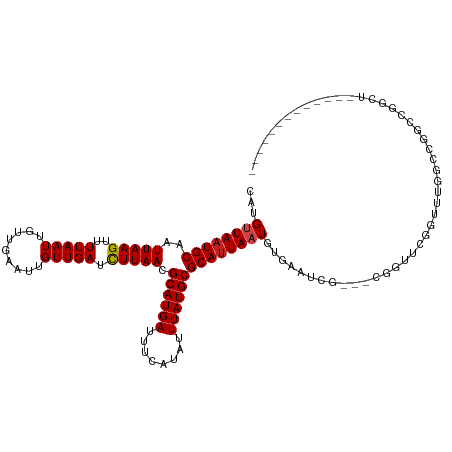
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:13 2011