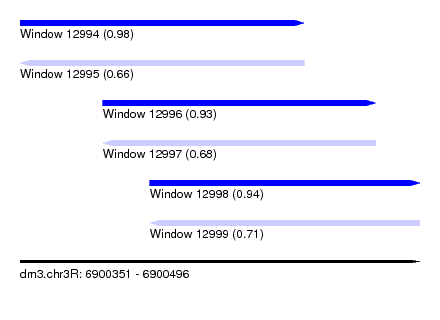
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,900,351 – 6,900,496 |
| Length | 145 |
| Max. P | 0.983907 |

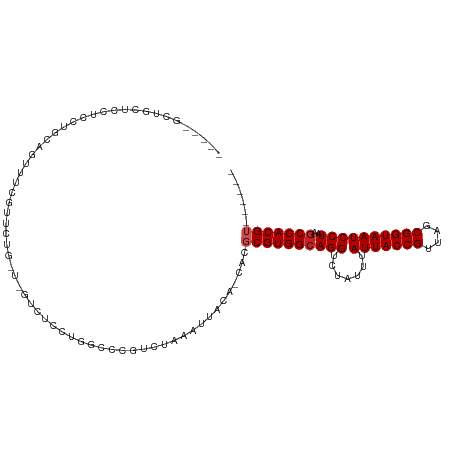
| Location | 6,900,351 – 6,900,454 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.48001 |
| G+C content | 0.51212 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6900351 103 + 27905053 ---CGGCUGCUCCUCUUGCAGUUUCGUCCUG-U-GUCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ ---.((((((.......))))))......((-(-((....(((.....)))....)))-)).((((((((((.......(((((((....))))))))))..)))))))------ ( -30.11, z-score = -2.00, R) >droSim1.chr3R 13049734 103 - 27517382 ---CGGCUGCUCCUCCUGCAGUUUCGUCCUG-U-GUCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ ---.((((((.......))))))......((-(-((....(((.....)))....)))-)).((((((((((.......(((((((....))))))))))..)))))))------ ( -30.11, z-score = -1.95, R) >droSec1.super_0 6056941 103 - 21120651 ---UGGCUGCUCCUCCUGCAGUUUCGUCCUG-U-GUCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ ---.((((((.......))))))......((-(-((....(((.....)))....)))-)).((((((((((.......(((((((....))))))))))..)))))))------ ( -30.01, z-score = -1.88, R) >droYak2.chr3R 10930145 107 + 28832112 CGGCUGCUCCUCCUCCUGCAGUUUCGUCGGG-UUGUCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ .((((.........((((((((((.(.((((-((.......)))))).).))))).((-(....)))))))).......(((((((....)))))))....))))....------ ( -35.70, z-score = -2.82, R) >droEre2.scaffold_4770 3029580 103 - 17746568 ---CCACUGCUCCUCCUGCAAUUUCGUCCUG-U-GUCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ ---....(((.......))).........((-(-((....(((.....)))....)))-)).((((((((((.......(((((((....))))))))))..)))))))------ ( -25.51, z-score = -1.79, R) >dp4.chr2 28377483 110 + 30794189 -----GCUGUUACUCUAUCUAUCUCCCUCUGAUACUGUUCUGUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGU -----(((((.((..((((...........))))..)).................))))).(((((((((((.......(((((((....))))))))))..)))))).)).... ( -28.88, z-score = -2.29, R) >droPer1.super_6 3715052 110 + 6141320 -----GCUGUUACUCUAUCUAUCUCCCUCUGAUACUCUUCUGUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGU -----(((((.........((((.......))))(......).............))))).(((((((((((.......(((((((....))))))))))..)))))).)).... ( -27.41, z-score = -2.23, R) >droVir3.scaffold_13047 14225216 100 - 19223366 ----CGCGUUGUCGGUGCCGUGUUGGUUCUG----UUCUGAGUUCCCUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU------ ----.((((((((((.(((.....))).)))----....(((....)))......)))-.))))((((((((.......(((((((....))))))))))..)))))..------ ( -29.91, z-score = -1.60, R) >droGri2.scaffold_14906 3766643 96 - 14172833 ---------CAUUCUCUCCGUACUGGUUGCG---ACUUUGAGUCCCCUCGAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGA------ ---------.........((((.....))))---..((((((....))))))......-....(((((((((.......(((((((....))))))))))..)))))).------ ( -25.31, z-score = -1.45, R) >droAna3.scaffold_13340 11461094 94 - 23697760 ----------UGCAGCUCCCUGCCCUCUCUG----UCUUUUGGCCCGGUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAUCCACGU------ ----------.((((....))))......((----(..((((((...))))))..)))-...((((((.(((.......(((((((....))))))))))...))))))------ ( -23.61, z-score = -1.14, R) >droMoj3.scaffold_6540 21429982 93 + 34148556 -----CCAGCGCCCGCCCCCUGGUGGCUG----------GAGUCCCCUUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGA------ -----(((((...((((....))))))))----------)..................-....(((((((((.......(((((((....))))))))))..)))))).------ ( -30.71, z-score = -1.60, R) >consensus _____GCUGCUCCUCCUGCAGUUUCGUUCUG_U_GUCUCCUGGCCCGUCUAAAUUACA_CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGU______ ..............................................................((((((((((.......(((((((....))))))))))..)))))))...... (-19.39 = -19.66 + 0.27)

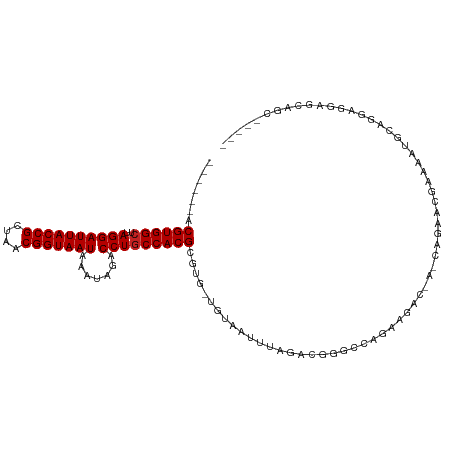
| Location | 6,900,351 – 6,900,454 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.48001 |
| G+C content | 0.51212 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6900351 103 - 27905053 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGAC-A-CAGGACGAAACUGCAAGAGGAGCAGCCG--- ------.((((((..((((((((((....))))))).......)))))))))(((.-((.....)).)))(((..(....)-.-(((.......)))...........))).--- ( -29.11, z-score = -1.10, R) >droSim1.chr3R 13049734 103 + 27517382 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGAC-A-CAGGACGAAACUGCAGGAGGAGCAGCCG--- ------.((((((..((((((((((....))))))).......)))))))))(((.-((.....)).)))(((..(....)-.-(((.......)))...........))).--- ( -29.11, z-score = -0.83, R) >droSec1.super_0 6056941 103 + 21120651 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGAC-A-CAGGACGAAACUGCAGGAGGAGCAGCCA--- ------.((((((..((((((((((....))))))).......)))))))))(((.-((.....)).)))(((..(....)-.-(((.......)))...........))).--- ( -29.11, z-score = -0.84, R) >droYak2.chr3R 10930145 107 - 28832112 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGACAA-CCCGACGAAACUGCAGGAGGAGGAGCAGCCG ------.((((((..((((((((((....))))))).......)))))))))....-((((.(((...((((...(....)..-))))...))).))))((..(.....)..)). ( -30.71, z-score = -0.74, R) >droEre2.scaffold_4770 3029580 103 + 17746568 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGAC-A-CAGGACGAAAUUGCAGGAGGAGCAGUGG--- ------..(((((..((((((((((....))))))).......))))))))((.(.-((((((((...((..((.(....)-.-..)).)))))))))).)....)).....--- ( -28.81, z-score = -1.11, R) >dp4.chr2 28377483 110 - 30794189 ACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAUUUAGACGAGACAGAACAGUAUCAGAGGGAGAUAGAUAGAGUAACAGC----- ....((.((((((..((((((((((....))))))).......))))))))))).(((((.((((...(......).....((((.......))))....)))).)))))----- ( -26.61, z-score = -1.20, R) >droPer1.super_6 3715052 110 - 6141320 ACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAUUUAGACGAGACAGAAGAGUAUCAGAGGGAGAUAGAUAGAGUAACAGC----- ....((.((((((..((((((((((....))))))).......))))))))))).(((((.((((...(......).....((((.......))))....)))).)))))----- ( -26.61, z-score = -1.25, R) >droVir3.scaffold_13047 14225216 100 + 19223366 ------ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGAGGGAACUCAGAA----CAGAACCAACACGGCACCGACAACGCG---- ------..(((((..((((((((((....))))))).......))))))))((((.-(((......(((....)))....----...........((....))))))))).---- ( -30.11, z-score = -2.56, R) >droGri2.scaffold_14906 3766643 96 + 14172833 ------UCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUCGAGGGGACUCAAAGU---CGCAACCAGUACGGAGAGAAUG--------- ------.((((((..((((((((((....))))))).......)))))))))((((-((..((((.(((....))).))))---..)).....)))).........--------- ( -29.41, z-score = -1.80, R) >droAna3.scaffold_13340 11461094 94 + 23697760 ------ACGUGGAUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAACCGGGCCAAAAGA----CAGAGAGGGCAGGGAGCUGCA---------- ------..(..(.(((..(((((((....)))))))...))).((((((..((.((-.((......)))).)).......----(.....)))))))...)..).---------- ( -25.30, z-score = -0.51, R) >droMoj3.scaffold_6540 21429982 93 - 34148556 ------UCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAAAGGGGACUC----------CAGCCACCAGGGGGCGGGCGCUGG----- ------..(((((..((((((((((....))))))).......))))))))((((.-..........((....))(----------(.(((.(....))))))))))...----- ( -31.11, z-score = -0.32, R) >consensus ______ACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG_UGUAAUUUAGACGGGCCAGAAGAC_A_CAGAACGAAAAUGCAGGAGGAGCAGC_____ .......((((((..((((((((((....))))))).......)))))))))............................................................... (-17.49 = -17.58 + 0.09)

| Location | 6,900,381 – 6,900,480 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Shannon entropy | 0.37572 |
| G+C content | 0.49530 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6900381 99 + 27905053 UCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUUGG--------------AAGACAAAAGGCAA .......(((.((((.(((..(.-(.(((((((((((.......(((((((....))))))))))..)))))).))).)..)))..--------------.))))....))).. ( -29.81, z-score = -1.21, R) >droSim1.chr3R 13049764 99 - 27517382 UCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUCGG--------------AAGACAAAAGGCAA ...(((.((((((.......)).-..(((((((((((.......(((((((....))))))))))..)))))).))).))).((..--------------..))....)))... ( -29.41, z-score = -1.15, R) >droSec1.super_0 6056971 99 - 21120651 UCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUCGG--------------AAGACAAAAGGCAA ...(((.((((((.......)).-..(((((((((((.......(((((((....))))))))))..)))))).))).))).((..--------------..))....)))... ( -29.41, z-score = -1.15, R) >droYak2.chr3R 10930179 99 + 28832112 UCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUCAG--------------AAGACAAAAGGCAG (((.((((.(((((.........-...((((((((((.......(((((((....))))))))))..))))))).)))))..))))--------------.))).......... ( -30.03, z-score = -2.00, R) >droEre2.scaffold_4770 3029610 99 - 17746568 UCUCCUGGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUCAG--------------AAGACAAAAGGCAG (((.((((.(((((.........-...((((((((((.......(((((((....))))))))))..))))))).)))))..))))--------------.))).......... ( -30.03, z-score = -2.00, R) >dp4.chr2 28377513 100 + 30794189 UGUUCUGUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCGGCGGCAUCA--------AAAGACAAA------ ...........((((........((.(((((((((((.......(((((((....))))))))))..)))))).))))((.....))....--------..))))...------ ( -30.21, z-score = -2.13, R) >droPer1.super_6 3715082 100 + 6141320 UCUUCUGUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCGGCGGCAUCA--------AAAGACAAA------ ...........((((........((.(((((((((((.......(((((((....))))))))))..)))))).))))((.....))....--------..))))...------ ( -30.21, z-score = -2.57, R) >droVir3.scaffold_13047 14225250 85 - 19223366 -------UUCCCUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAA-GU--------------GAAACAAA------ -------...........((((.-(.(((((((((((.......(((((((....))))))))))..)))))).))..)....-))--------------))......------ ( -22.51, z-score = -1.50, R) >droGri2.scaffold_14906 3766673 84 - 14172833 -------UCCCCUCGAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGAUAGCAGCA--CU--------------GAAACAAA------ -------................-....(((((((((.......(((((((....))))))))))..)))))).........--..--------------........------ ( -19.51, z-score = -1.53, R) >droAna3.scaffold_13340 11461115 99 - 23697760 UCUUUUGGCCCGGUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAUCCACGUCGGCGGCAUCAA--------------AAGACAAAAGGCAC ((((((((.(((((......)).-..(((((((.(((.......(((((((....))))))))))...))))).)).)))..))))--------------)))).......... ( -28.21, z-score = -2.09, R) >droWil1.scaffold_181130 7632849 112 + 16660200 -UCGUCGCCUCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACAGUUAACGGUAAUCCUAAGCCACGACGGCGGCGGUGCCAACGUCAGCAUCAAAGACAAAAGGCAA -.....((((.((((........-..(((((((((((.......(((((.(....).))))))))..)))))).))((((((.......)))).)).....))))...)))).. ( -32.21, z-score = -1.80, R) >droMoj3.scaffold_6540 21430009 86 + 34148556 -------UCCCCUUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGACGGCGCCGAGCU--------------GAAACAAA------ -------................-....(((((((((.......(((((((....))))))))))..)))))).((((.....)))--------------).......------ ( -24.61, z-score = -1.96, R) >consensus UCUCCUGGCCCGUCUAAAUUACA_CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAUCGG______________AAGACAAAAGGCA_ ..........................(((((((((((.......(((((((....))))))))))..)))))).))...................................... (-20.54 = -20.72 + 0.17)


| Location | 6,900,381 – 6,900,480 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Shannon entropy | 0.37572 |
| G+C content | 0.49530 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6900381 99 - 27905053 UUGCCUUUUGUCUU--------------CCAAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGA ..((((...((((.--------------..(((..(((((.((((((..((((((((((....))))))).......))))))))))).)-))..))).))))))))....... ( -32.81, z-score = -1.89, R) >droSim1.chr3R 13049764 99 + 27517382 UUGCCUUUUGUCUU--------------CCGAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGA ..((((...((((.--------------..(((..(((((.((((((..((((((((((....))))))).......))))))))))).)-))..))).))))))))....... ( -33.21, z-score = -1.86, R) >droSec1.super_0 6056971 99 + 21120651 UUGCCUUUUGUCUU--------------CCGAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGA ..((((...((((.--------------..(((..(((((.((((((..((((((((((....))))))).......))))))))))).)-))..))).))))))))....... ( -33.21, z-score = -1.86, R) >droYak2.chr3R 10930179 99 - 28832112 CUGCCUUUUGUCUU--------------CUGAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGA ((((((...((((.--------------..(((..(((((.((((((..((((((((((....))))))).......))))))))))).)-))..))).))))))).))).... ( -34.81, z-score = -2.48, R) >droEre2.scaffold_4770 3029610 99 + 17746568 CUGCCUUUUGUCUU--------------CUGAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGGGCCAGGAGA ((((((...((((.--------------..(((..(((((.((((((..((((((((((....))))))).......))))))))))).)-))..))).))))))).))).... ( -34.81, z-score = -2.48, R) >dp4.chr2 28377513 100 - 30794189 ------UUUGUCUUU--------UGAUGCCGCCGACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAUUUAGACGAGACAGAACA ------((((((((.--------...........((((((.((((((..((((((((((....))))))).......))))))))))).)).)).........))))))))... ( -30.01, z-score = -2.33, R) >droPer1.super_6 3715082 100 - 6141320 ------UUUGUCUUU--------UGAUGCCGCCGACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAUUUAGACGAGACAGAAGA ------((((((((.--------...........((((((.((((((..((((((((((....))))))).......))))))))))).)).)).........))))))))... ( -30.01, z-score = -2.30, R) >droVir3.scaffold_13047 14225250 85 + 19223366 ------UUUGUUUC--------------AC-UUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGAGGGAA------- ------....(..(--------------.(-(...(((((.((((((..((((((((((....))))))).......))))))))))).)-))......)).)..).------- ( -24.31, z-score = -1.13, R) >droGri2.scaffold_14906 3766673 84 + 14172833 ------UUUGUUUC--------------AG--UGCUGCUAUCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUCGAGGGGA------- ------.((.((((--------------..--(((.((...((((((..((((((((((....))))))).......)))))))))...)-)))).....)))).))------- ( -21.01, z-score = 0.13, R) >droAna3.scaffold_13340 11461115 99 + 23697760 GUGCCUUUUGUCUU--------------UUGAUGCCGCCGACGUGGAUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAACCGGGCCAAAAGA ....(((((((((.--------------..(((..(((((.(((((..(((((((((((....))))))).......))))))))))).)-))..))).....))).)))))). ( -28.21, z-score = -1.38, R) >droWil1.scaffold_181130 7632849 112 - 16660200 UUGCCUUUUGUCUUUGAUGCUGACGUUGGCACCGCCGCCGUCGUGGCUUAGGAUUACCGUUAACUGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAGACGAGGCGACGA- (((((((..((((..(.(((..(((((((((..(((((....)))))(((..(((((.(....).)))))...)))...))))).)))).-.))).)..)))))))))))...- ( -35.30, z-score = -1.66, R) >droMoj3.scaffold_6540 21430009 86 - 34148556 ------UUUGUUUC--------------AGCUCGGCGCCGUCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAUUUAAAGGGGA------- ------........--------------..(((.((((((.((((((..((((((((((....))))))).......))))))))))).)-)))........)))..------- ( -26.21, z-score = -0.75, R) >consensus _UGCCUUUUGUCUU______________CCGAUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG_UGUAAUUUAGACGGGCCAGGAGA ......................................((.((((((..((((((((((....))))))).......))))))))))).......................... (-18.56 = -18.74 + 0.17)

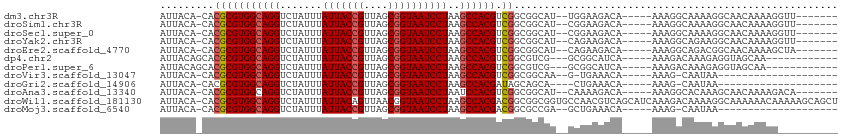
| Location | 6,900,398 – 6,900,496 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.38941 |
| G+C content | 0.47357 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
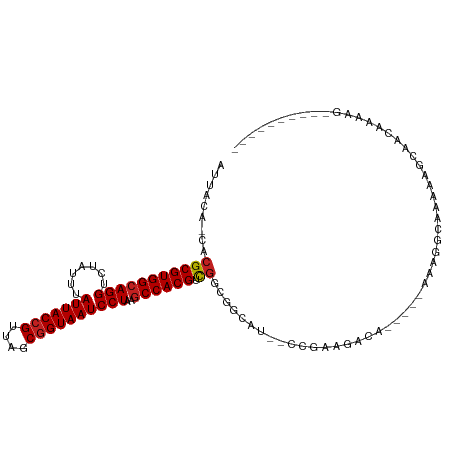
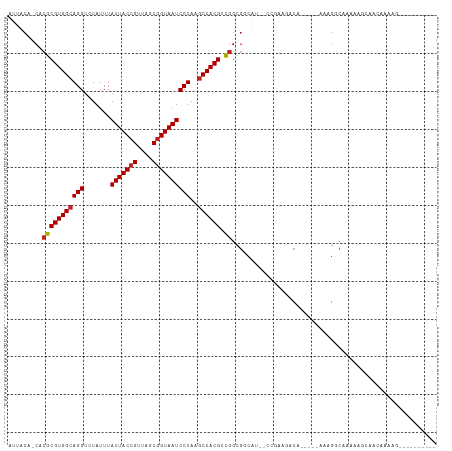
>dm3.chr3R 6900398 98 + 27905053 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU--UGGAAGACA-----AAAGGCAAAAGGCAACAAAAGGUU------- ......-..(((((((((((.......(((((((....))))))))))..)))))).))...((.(--((.....))-----)...))....(....)........------- ( -25.81, z-score = -1.34, R) >droSim1.chr3R 13049781 98 - 27517382 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU--CGGAAGACA-----AAAGGCAAAAGGCAACAAAAGGUU------- ......-..(((((((((((.......(((((((....))))))))))..)))))).))((....(--(....))..-----....))....(....)........------- ( -26.11, z-score = -1.57, R) >droSec1.super_0 6056988 98 - 21120651 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU--CGGAAGACA-----AAAGGCAAAAGGCAACAAAAGGUU------- ......-..(((((((((((.......(((((((....))))))))))..)))))).))((....(--(....))..-----....))....(....)........------- ( -26.11, z-score = -1.57, R) >droYak2.chr3R 10930196 98 + 28832112 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU--CAGAAGACA-----AAAGGCAGAAGGCAACAAAAGGUU------- ......-...((((((((((.......(((((((....))))))))))..)))))(((..(.....--..)..))).-----....))....(....)........------- ( -24.91, z-score = -1.44, R) >droEre2.scaffold_4770 3029627 98 - 17746568 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU--CAGAAGACA-----AAAGGCAGACGGCAACAAAAGCUA------- ......-...((((((((((.......(((((((....))))))))))..)))))((((...((..--.........-----....))...)))).......))..------- ( -26.47, z-score = -1.46, R) >dp4.chr2 28377530 93 + 30794189 AUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCG---GCGGCAUCA-----AAAGACAAAGAGGUAGCAA------------ .((((.((.(((((((((((.......(((((((....))))))))))..)))))).))))(((.---.........-----...)))......))))...------------ ( -28.53, z-score = -1.81, R) >droPer1.super_6 3715099 93 + 6141320 AUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCG---GCGGCAUCA-----AAAGACAAAGAGGUAGCAA------------ .((((.((.(((((((((((.......(((((((....))))))))))..)))))).))))(((.---.........-----...)))......))))...------------ ( -28.53, z-score = -1.81, R) >droVir3.scaffold_13047 14225260 83 - 19223366 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAA--G-UGAAACA-----AAAG-CAAUAA-------------------- .((((.-(.(((((((((((.......(((((((....))))))))))..)))))).))..)....--)-)))....-----....-......-------------------- ( -22.51, z-score = -1.50, R) >droGri2.scaffold_14906 3766683 82 - 14172833 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGAUAGCAGCA----CUGAAACA-----AAAG-CAAUAA-------------------- ......-...((((((((((.......(((((((....))))))))))..)))))..(((.....----))).....-----...)-).....-------------------- ( -19.71, z-score = -1.73, R) >droAna3.scaffold_13340 11461132 98 - 23697760 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAUCCACGUCGGCGGCAU--CAAAAGACA-----AAAGGCACAAAGCAACAAAAGACA------- ......-..(((((((.(((.......(((((((....))))))))))...))))).))((.((..--.........-----....)).....))...........------- ( -19.67, z-score = -0.83, R) >droWil1.scaffold_181130 7632865 112 + 16660200 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACAGUUAACGGUAAUCCUAAGCCACGACGGCGGCGGUGCCAACGUCAGCAUCAAAGACAAAAGGCAAAAAACAAAAAGCAGCU ......-..(((((((((((.......(((((.(....).))))))))..)))))).))((.(((((((........)))))..........(........).....)).)). ( -25.01, z-score = -0.52, R) >droMoj3.scaffold_6540 21430019 84 + 34148556 AUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGACGGCGCCGA--GCUGAAACA-----AAAG-CAAUAA-------------------- ......-...((((((((((.......(((((((....))))))))))..)))))..((((.....--)))).....-----...)-).....-------------------- ( -25.61, z-score = -2.23, R) >consensus AUUACA_CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGGCAU__CCGAAGACA_____AAAGGCAAAAAGCAACAAAAG__________ .........(((((((((((.......(((((((....))))))))))..)))))).))...................................................... (-20.54 = -20.72 + 0.17)
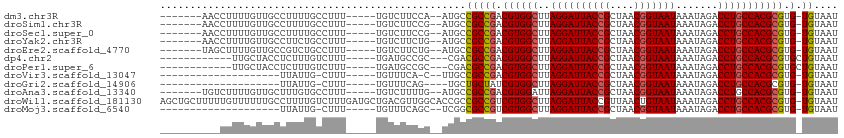
| Location | 6,900,398 – 6,900,496 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.38941 |
| G+C content | 0.47357 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6900398 98 - 27905053 -------AACCUUUUGUUGCCUUUUGCCUUU-----UGUCUUCCA--AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------.....(((((((((.((.((....-----.((..(((.--..(((((....)))))...)))..)).)).)).)))))))))((.((.(((...))).)-).)).. ( -23.90, z-score = -1.21, R) >droSim1.chr3R 13049781 98 + 27517382 -------AACCUUUUGUUGCCUUUUGCCUUU-----UGUCUUCCG--AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------.....(((((((((.((.((....-----.((..(((.--..(((((....)))))...)))..)).)).)).)))))))))((.((.(((...))).)-).)).. ( -25.00, z-score = -1.46, R) >droSec1.super_0 6056988 98 + 21120651 -------AACCUUUUGUUGCCUUUUGCCUUU-----UGUCUUCCG--AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------.....(((((((((.((.((....-----.((..(((.--..(((((....)))))...)))..)).)).)).)))))))))((.((.(((...))).)-).)).. ( -25.00, z-score = -1.46, R) >droYak2.chr3R 10930196 98 - 28832112 -------AACCUUUUGUUGCCUUCUGCCUUU-----UGUCUUCUG--AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------........................-----.(((....)--))..(((((.((((((..((((((((((....))))))).......))))))))))).)-)).... ( -24.01, z-score = -1.25, R) >droEre2.scaffold_4770 3029627 98 + 17746568 -------UAGCUUUUGUUGCCGUCUGCCUUU-----UGUCUUCUG--AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------.....(((((((((((..((....-----.(((..(((--(.(((((....))))))))))))....))..)))))))))))((.((.(((...))).)-).)).. ( -28.70, z-score = -1.73, R) >dp4.chr2 28377530 93 - 30794189 ------------UUGCUACCUCUUUGUCUUU-----UGAUGCCGC---CGACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAU ------------((((.........(((...-----.........---.)))((((.((((((..((((((((((....))))))).......))))))))))).)).)))). ( -25.53, z-score = -1.31, R) >droPer1.super_6 3715099 93 - 6141320 ------------UUGCUACCUCUUUGUCUUU-----UGAUGCCGC---CGACGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUGCUGUAAU ------------((((.........(((...-----.........---.)))((((.((((((..((((((((((....))))))).......))))))))))).)).)))). ( -25.53, z-score = -1.31, R) >droVir3.scaffold_13047 14225260 83 + 19223366 --------------------UUAUUG-CUUU-----UGUUUCA-C--UUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU --------------------......-....-----.......-.--....(((((.((((((..((((((((((....))))))).......))))))))))).)-)).... ( -23.41, z-score = -1.55, R) >droGri2.scaffold_14906 3766683 82 + 14172833 --------------------UUAUUG-CUUU-----UGUUUCAG----UGCUGCUAUCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU --------------------......-....-----........----(((.((...((((((..((((((((((....))))))).......)))))))))...)-)))).. ( -19.91, z-score = -0.43, R) >droAna3.scaffold_13340 11461132 98 + 23697760 -------UGUCUUUUGUUGCUUUGUGCCUUU-----UGUCUUUUG--AUGCCGCCGACGUGGAUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU -------...........((.....))....-----.(((....)--))..(((((.(((((..(((((((((((....))))))).......))))))))))).)-)).... ( -22.21, z-score = -0.30, R) >droWil1.scaffold_181130 7632865 112 - 16660200 AGCUGCUUUUUGUUUUUUGCCUUUUGUCUUUGAUGCUGACGUUGGCACCGCCGCCGUCGUGGCUUAGGAUUACCGUUAACUGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU .................((((...((((.........))))..))))....(((((.((((((..((((((((.(....).))))).......))))))))))).)-)).... ( -27.11, z-score = -0.20, R) >droMoj3.scaffold_6540 21430019 84 - 34148556 --------------------UUAUUG-CUUU-----UGUUUCAGC--UCGGCGCCGUCGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG-UGUAAU --------------------.....(-((..-----......)))--...((((((.((((((..((((((((((....))))))).......))))))))))).)-)))... ( -26.81, z-score = -1.69, R) >consensus __________CUUUUGUUGCCUUUUGCCUUU_____UGUCUUCGG__AUGCCGCCGACGUGGCUUAGGAUUACCGCUAACGGUAAUAAAUAGACCUGCCACGCGUG_UGUAAU ......................................................((.((((((..((((((((((....))))))).......)))))))))))......... (-18.56 = -18.74 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:10 2011