| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,883,865 – 6,883,963 |
| Length | 98 |
| Max. P | 0.775377 |

| Location | 6,883,865 – 6,883,963 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Shannon entropy | 0.10597 |
| G+C content | 0.43473 |
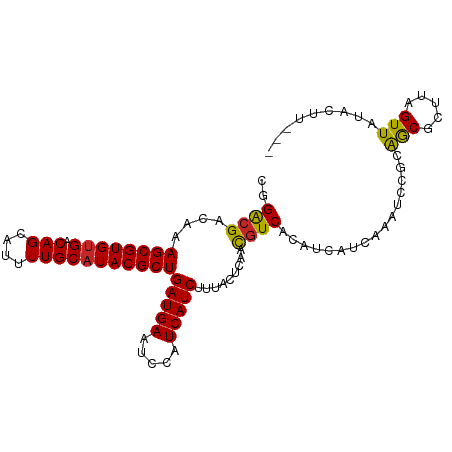
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -20.77 |
| Energy contribution | -20.49 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
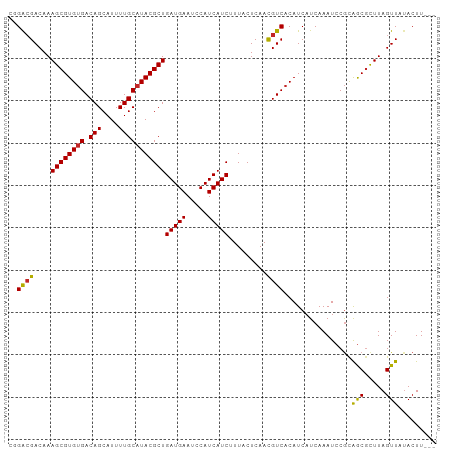
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
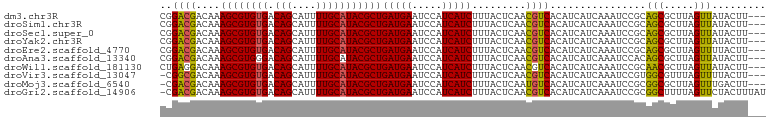
>dm3.chr3R 6883865 98 + 27905053 CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUAUACUU--- ..((((....((((((((.(((....)))))))))))(((((.....))))).........))))................(((.....)))......--- ( -21.80, z-score = -1.25, R) >droSim1.chr3R 13038871 98 - 27517382 CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUAUACUU--- ..((((....((((((((.(((....)))))))))))(((((.....))))).........))))................(((.....)))......--- ( -21.80, z-score = -1.25, R) >droSec1.super_0 6046061 98 - 21120651 CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUAUACUU--- ..((((....((((((((.(((....)))))))))))(((((.....))))).........))))................(((.....)))......--- ( -21.80, z-score = -1.25, R) >droYak2.chr3R 10919081 98 + 28832112 CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUAUACUU--- ..((((....((((((((.(((....)))))))))))(((((.....))))).........))))................(((.....)))......--- ( -21.80, z-score = -1.25, R) >droEre2.scaffold_4770 3018796 98 - 17746568 CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUUUACUU--- ..((((....((((((((.(((....)))))))))))(((((.....))))).........)))).................................--- ( -21.60, z-score = -1.20, R) >droAna3.scaffold_13340 11450592 98 - 23697760 CGGACGACAAAGCGUGGGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCACAGCGCUUAGUUAUACUU--- .....(((.((((((.(((..((.....))....(.(((((....))))).)........................)))...)))))).)))......--- ( -19.90, z-score = -1.15, R) >droWil1.scaffold_181130 7612023 98 + 16660200 CUGAGGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAACGCUUAGUUAUACUU--- .(((.(((..((((((((.(((....)))))))))))(((((.....)))))..........)))...)))..........(((.....)))......--- ( -21.60, z-score = -1.57, R) >droVir3.scaffold_13047 14213935 97 - 19223366 -CGGCGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGUGGCGUUUAGUUUUACUU--- -.(....)..((((((((.(((....)))))))))))(((((.....)))))...(((.((((((((............)))))))).))).......--- ( -27.20, z-score = -2.82, R) >droMoj3.scaffold_6540 21418545 97 + 34148556 -CGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAAUGUCACAUCAUCAAAUCCGCGGCGCUUAGUUUGACUU--- -....((((.((((((((.(((....)))))))))))(((((.....))))).........))))......((((((..((...))...))))))...--- ( -23.30, z-score = -1.05, R) >droGri2.scaffold_14906 3755088 100 - 14172833 -CGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCGGCUUUUAGUUCUACUUUAU -.((((....((((((((.(((....)))))))))))(((((.....))))).........)))).................................... ( -21.90, z-score = -1.82, R) >consensus CGGACGACAAAGCGUGUGACAGCAUUUUGCAUACGCUGAUGAAUCCAUCAUCUUUACUCAACGUCACAUCAUCAAAUCCGCAGCGCUUAGUUAUACUU___ ..((((....((((((((.(((....)))))))))))(((((.....))))).........))))................(((.....)))......... (-20.77 = -20.49 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:04 2011