
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,951,746 – 6,951,858 |
| Length | 112 |
| Max. P | 0.744119 |

| Location | 6,951,746 – 6,951,858 |
|---|---|
| Length | 112 |
| Sequences | 14 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.19 |
| Shannon entropy | 0.40013 |
| G+C content | 0.43176 |
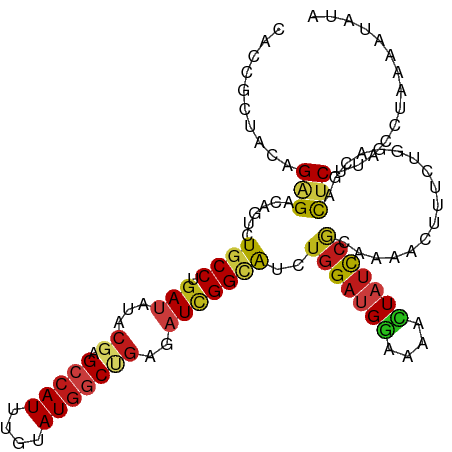
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.31 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
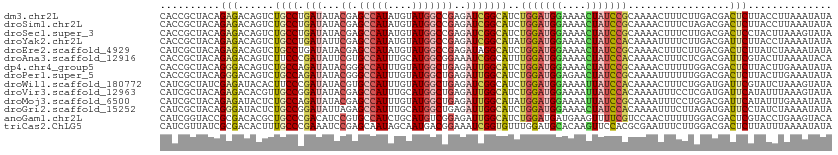
>dm3.chr2L 6951746 112 + 23011544 CACCGCUACAGAGACAGUCUGCCUGAUAUACGAGCCAUAUGUAUGGCCGAGAUCGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUCUUGACGACUCUUACCUUAAAUAUA .........((((...(((((((.(((...((.(((((....)))))))..)))))))...((((((....)))))).............)))..))))............. ( -29.50, z-score = -2.71, R) >droSim1.chr2L 6748651 112 + 22036055 CACCGCUACAGAGACAGUCUGCCUGAUAUACGAGCCAUAUGUAUGGCCGAGAUCGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUCUAGACGACUCUUACCUUAAAUAUA .........((((...(((((((.(((...((.(((((....)))))))..))))))....((((((....))))))............))))..))))............. ( -30.20, z-score = -2.83, R) >droSec1.super_3 2484438 112 + 7220098 CACCGCUACAGAGACAGUCUGCCUGAUAUACGAGCCAUAUGUAUGGCCGAGAUCGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUCUUGACGACUCCUACUUAAAGUAUA ..........(((...(((((((.(((...((.(((((....)))))))..)))))))...((((((....)))))).............)))..))).............. ( -28.50, z-score = -1.88, R) >droYak2.chr2L 16366669 112 - 22324452 CACCGCUACAGAGACAGUCUGCCUGAUAUUCGAGCCAUAUGUAUGGCCGAGAUCGGCAUAUGGAUGGAAAACUAUCCACAAAAUUUUCUUGACGAUUCUUACCUAAAAUAUA ..........((((..(((((((.(((.((((.(((((....))))))))))))))))..(((((((....)))))))............)))...))))............ ( -31.70, z-score = -3.59, R) >droEre2.scaffold_4929 15867708 112 + 26641161 CAUCGCUACAGAGACAGUCUGCCUGAUAUACGAGCCAUAUGUAUGGCCGAGAUAGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUCUUGACGACUCUUAUCUAAAAUAUA .........((((...(((((((((...(.((.(((((....))))))))..))))))...((((((....)))))).............)))..))))............. ( -28.40, z-score = -2.06, R) >droAna3.scaffold_12916 8535108 112 + 16180835 CACCGCUACAGAGACAGUCUUCCCGAUAUUCGUGCCAUUUGCAUGGCGGAAAUCGGCAUUUGGAUGGAAAACUAUCCACAAAACUUUCUCGACGAUUCGUACUUAAAAUACA ..........((((.(((....(((((.(((.((((((....))))))))))))))....(((((((....)))))))....))).))))...................... ( -28.80, z-score = -2.34, R) >dp4.chr4_group5 448453 112 + 2436548 CACCGCUACAGGGACAGUCUGCCAGAUAUACGGGCCAUUUGUAUGGCUGAGAUUGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUUUGGACGACUCUUACUUGAAAUAUA ........((((...((((..(((((......((((((....)))))).((((....))))((((((....)))))).........)))))..))))....))))....... ( -28.50, z-score = -1.42, R) >droPer1.super_5 4946478 112 + 6813705 CACCGCUACAGGGACAGUCUGCCAGAUAUACGGGCCAUUUGUAUGGCUGAGAUUGGCAUCUGGAUGGAGAACUAUCCGCAAAAUUUUUUGGACGACUCUUACUUGAAAUAUA ........((((...((((..(((((......((((((....)))))).((((....))))((((((....)))))).........)))))..))))....))))....... ( -28.50, z-score = -1.12, R) >droWil1.scaffold_180772 2156499 112 - 8906247 CAUCGCUAUCGAGAUACACUUCCCGAUAUACGUGCCAUUUGUAUGGCUGAGAUCGGCAUCUGGAUGGAAAAUUAUCCACAAAACUUUCUGGAUGAUUCGUAUCUAAAGUAUA ....(((....((((((.....(((((...((.(((((....)))))))..)))))((((..((((((......))))........))..))))....))))))..)))... ( -31.60, z-score = -2.52, R) >droVir3.scaffold_12963 5490940 112 - 20206255 CAUCGCUACAGAGACACGUUGCCGGAUAUACGAGCCAUUUGCAUGGCUGAGAUUGGCAUCUGGAUGGAAAAUUAUCCACAAAAUUUCCUCGAUGAUUCAUAUUUAAAGUAUA (((((...((((.......(((((......).((((((....))))))......))))))))..((((......))))...........))))).................. ( -22.81, z-score = 0.16, R) >droMoj3.scaffold_6500 14697438 112 - 32352404 CAUCGCUACAGAGAUACUCUGCCAGAUAUACGAGCCAUUUGUAUGGCUGAGAUUGGCAUAUGGAUGGAAAAUUAUCCGCAAAAUUUCCUGGACGAUUCAUAUUUGAAAUAUA ..(((...(((.((.....(((..((((......(((((..((((.(.......).))))..))))).....)))).))).....)))))..)))((((....))))..... ( -27.50, z-score = -1.54, R) >droGri2.scaffold_15252 7899608 112 - 17193109 CAUCGCUACAGGGAUACUCUGCCGGAUAUUAGAGCCAUUUGCAUGGCUGAGAUUGGCAUCUGGAUGGAAAACUAUCCACAAAAUUUCUUAGAUGAUUCCUAUCUAAAAUAUA .........((((((..(((((((...(((..((((((....))))))..)))))))...(((((((....)))))))...........)))..))))))............ ( -27.70, z-score = -1.31, R) >anoGam1.chr2L 39974052 112 + 48795086 CAUCGGUACCGCGACACGCUGCCCGACAUCCGUGCCAUCUGCAUGUCGGAGAUUGGCAUCUGGAUGAUGAAGUUUUCGUCCAACUUUUUGGACGACUCGUACCUGAAGUACA ..((((....(((...)))...))))((((((((((((((.(.....).))).))))))..))))).........((((((((....))))))))...((((.....)))). ( -37.60, z-score = -1.83, R) >triCas2.ChLG5 10251087 112 + 18847211 CAUCGUUAUCGCGACACUUUGCCCGAAAUCCGAGCAAUAGCAAUGACGGAAAUCGGUGUUUGGAUGCACAAGUUCCACGCGAAUUUCUUGGACGACUCUUAUUUAAAAUAUA ..(((((.(((((.....((((.((.....)).))))..........((((....((((......))))...)))).)))))........)))))................. ( -24.80, z-score = -0.22, R) >consensus CACCGCUACAGAGACAGUCUGCCUGAUAUACGAGCCAUUUGUAUGGCUGAGAUCGGCAUCUGGAUGGAAAACUAUCCGCAAAACUUUCUGGACGACUCUUACCUAAAAUAUA ..........(((......((((.(((...((.(((((....)))))))..)))))))..(((((((....))))))).................))).............. (-16.28 = -16.31 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:36 2011