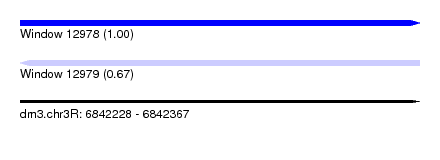
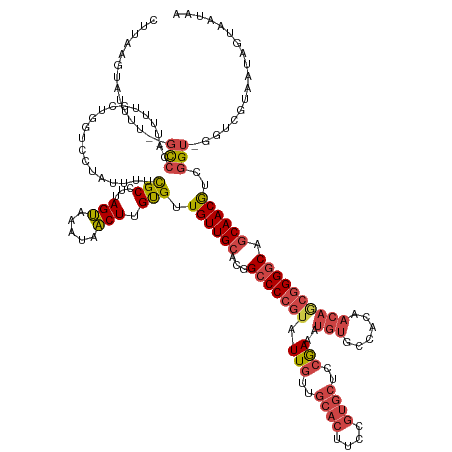
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,842,228 – 6,842,367 |
| Length | 139 |
| Max. P | 0.995689 |

| Location | 6,842,228 – 6,842,367 |
|---|---|
| Length | 139 |
| Sequences | 6 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 84.02 |
| Shannon entropy | 0.30261 |
| G+C content | 0.44233 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -29.12 |
| Energy contribution | -31.60 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
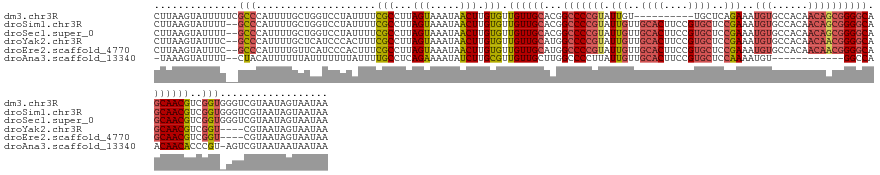
>dm3.chr3R 6842228 139 + 27905053 UUAUUACUAUUACGACCCACCGACGUUGCUGCCCCGCUGUUGUGGCACAUUUCUGAGCA----------ACAAUACGGGGCCGUGCAACAACACAAGUUAUUUACUAAGGCGAAAAUAGGACCAGCAAAAUGGGCGAAAAAUACUUAAG ............((.((((.....(((((.((((((.((((((.((.((....)).)).----------))))))))))))...)))))...................(((........).)).......))))))............. ( -37.80, z-score = -1.78, R) >droSim1.chr3R 12997243 147 - 27517382 UUAUUACUAUUACGACCCACCGACGUUGCUGCCCCGCUGUUGUGGCACAUUUCGGAGCACGGAAGUGCAACAAUACGGGGCCGUGCAACAACACAAGUUAUUUACUAAGGCGAAAAUAGGACCAGCAAAAUGGGC--AAAAUACUUAAG ......(((((.((.((.......(((((.((((((.((((((.((((.(((((.....))))))))).))))))))))))...)))))......(((.....)))..))))..)))))..(((......)))..--............ ( -46.70, z-score = -2.97, R) >droSec1.super_0 6004783 147 - 21120651 UUAUUACUAUUACGACCCACCGACGUUGCUGCCCCGCUGUUGUGGCACAUUUCGGAGCACGGAAGUGCAACAAUACGGGGCCGUGCAACAACACAAGUUAUUUACUAAGGCGAAAAUAGGACCAGCAAAAUGGGC--AAAAUACUUAAG ......(((((.((.((.......(((((.((((((.((((((.((((.(((((.....))))))))).))))))))))))...)))))......(((.....)))..))))..)))))..(((......)))..--............ ( -46.70, z-score = -2.97, R) >droYak2.chr3R 10876793 143 + 28832112 UUAUUACUAUUACG----ACCGACGUUGCUGCCCCGUUGUUGUGGCACAUUUCGGAGCACGGAAGUGCAACAAUACGGGGCCAUGCAACAACACAAGUUAUUUACUAAGGCGAAAGUGGGAUGAGCAAAAUGGGC--GAAAUACUUAAG .......((((.((----.((.(..((((((((((((.(((((.((((.(((((.....))))))))).))))))))))))(((.(.........(((.....)))...((....)).).))))))))..).)))--).))))...... ( -47.50, z-score = -3.53, R) >droEre2.scaffold_4770 2976493 143 - 17746568 UUAUUACUAUUACG----ACCGACGUUGCUGCCCCGUUGUUGUGGCACAUUUCGGAGCACGGAAGUGCAACAAUACGGGGCCAUGCAACAACACAAGUUAUUUACUAAGGCGAAAGUGGGAUGAACAAAAUGGGC--GAAAUACUUAAG .......((((.((----.((...(((((.(((((((.(((((.((((.(((((.....))))))))).))))))))))))...)))))....((..((.((((((...((....)).)).)))).))..)))))--).))))...... ( -47.20, z-score = -3.70, R) >droAna3.scaffold_13340 11411277 133 - 23697760 UUAUUAUUAUUACGACU-ACGGGUGUUGUUGGCC------------ACAUUUUGGAGCACGGAAGUGCAACAAUAAGGGGCCAAGCAACAACGCAAGAUAUUUUCUGAGGCAAAAUAAAAAAAUAAAAAAUGUAG--AAAAUACUUUA- ...............((-((((.(((((((((((------------.....(((..((((....))))..))).....)))).))))))).)((.(((.....)))...))...................)))))--...........- ( -37.30, z-score = -4.78, R) >consensus UUAUUACUAUUACGACC_ACCGACGUUGCUGCCCCGCUGUUGUGGCACAUUUCGGAGCACGGAAGUGCAACAAUACGGGGCCAUGCAACAACACAAGUUAUUUACUAAGGCGAAAAUAGGACCAGCAAAAUGGGC__AAAAUACUUAAG ......(((((.............(((((.((((((.((((((.((((.(((((.....))))))))).))))))))))))...)))))......(((.....)))........))))).............................. (-29.12 = -31.60 + 2.48)

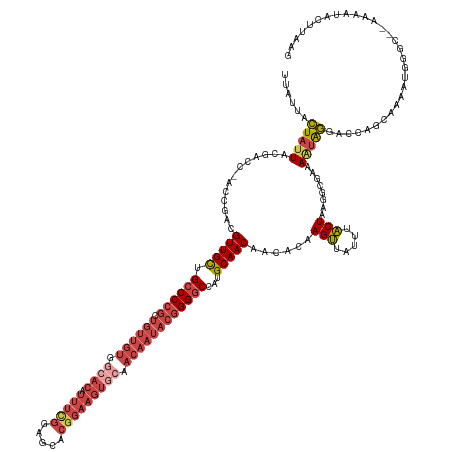
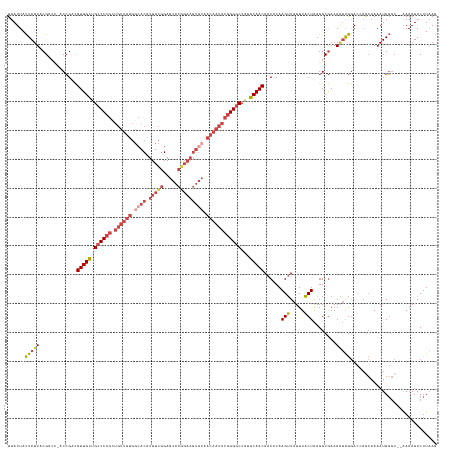
| Location | 6,842,228 – 6,842,367 |
|---|---|
| Length | 139 |
| Sequences | 6 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 84.02 |
| Shannon entropy | 0.30261 |
| G+C content | 0.44233 |
| Mean single sequence MFE | -40.24 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.68 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.669925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6842228 139 - 27905053 CUUAAGUAUUUUUCGCCCAUUUUGCUGGUCCUAUUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCACGGCCCCGUAUUGU----------UGCUCAGAAAUGUGCCACAACAGCGGGGCAGCAACGUCGGUGGGUCGUAAUAGUAAUAA ....(.((((...((((((((((((((((.........)))..)))))).......((.((((((...(((((((.((((----------.((.((....)).)).))))..))))))).)))))).))))))).)).)))).)..... ( -44.80, z-score = -2.39, R) >droSim1.chr3R 12997243 147 + 27517382 CUUAAGUAUUUU--GCCCAUUUUGCUGGUCCUAUUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCACGGCCCCGUAUUGUUGCACUUCCGUGCUCCGAAAUGUGCCACAACAGCGGGGCAGCAACGUCGGUGGGUCGUAAUAGUAAUAA ....(.((((..--(((((((((((((((.........)))..)))))).......((.((((((...(((((((.((((.((((((.((.....)).)).)))).))))..))))))).)))))).))))))))...)))).)..... ( -44.60, z-score = -1.25, R) >droSec1.super_0 6004783 147 + 21120651 CUUAAGUAUUUU--GCCCAUUUUGCUGGUCCUAUUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCACGGCCCCGUAUUGUUGCACUUCCGUGCUCCGAAAUGUGCCACAACAGCGGGGCAGCAACGUCGGUGGGUCGUAAUAGUAAUAA ....(.((((..--(((((((((((((((.........)))..)))))).......((.((((((...(((((((.((((.((((((.((.....)).)).)))).))))..))))))).)))))).))))))))...)))).)..... ( -44.60, z-score = -1.25, R) >droYak2.chr3R 10876793 143 - 28832112 CUUAAGUAUUUC--GCCCAUUUUGCUCAUCCCACUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCAUGGCCCCGUAUUGUUGCACUUCCGUGCUCCGAAAUGUGCCACAACAACGGGGCAGCAACGUCGGU----CGUAAUAGUAAUAA ....(.((((.(--(((.((.(((((..(((((((........)))...........(((((((..((((..(((.(((..((((....))))..))).))).))))))))))))))).))))).)).)).----)).)))).)..... ( -39.90, z-score = -2.11, R) >droEre2.scaffold_4770 2976493 143 + 17746568 CUUAAGUAUUUC--GCCCAUUUUGUUCAUCCCACUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCAUGGCCCCGUAUUGUUGCACUUCCGUGCUCCGAAAUGUGCCACAACAACGGGGCAGCAACGUCGGU----CGUAAUAGUAAUAA ....(.((((.(--(.((...................(((...(((.....))).)))..(((((...(((((((.((((.((((((.((.....)).)).)))).))))..))))))).)))))...)).----)).)))).)..... ( -37.60, z-score = -1.57, R) >droAna3.scaffold_13340 11411277 133 + 23697760 -UAAAGUAUUUU--CUACAUUUUUUAUUUUUUUAUUUUGCCUCAGAAAAUAUCUUGCGUUGUUGCUUGGCCCCUUAUUGUUGCACUUCCGUGCUCCAAAAUGU------------GGCCAACAACACCCGU-AGUCGUAAUAAUAAUAA -....(((((((--((...........................))))))))).(((((.(((((.((((((.(...(((..((((....))))..)))...).------------)))))))))))..)))-))............... ( -29.93, z-score = -3.50, R) >consensus CUUAAGUAUUUU__GCCCAUUUUGCUGGUCCUAUUUUCGCCUUAGUAAAUAACUUGUGUUGUUGCACGGCCCCGUAUUGUUGCACUUCCGUGCUCCGAAAUGUGCCACAACAGCGGGGCAGCAACGUCGGU_GGUCGUAAUAGUAAUAA ..............(((....................(((...(((.....))).))).((((((...(((((((.(((..((((....))))..)))..(((......)))))))))).))))))..))).................. (-24.44 = -25.68 + 1.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:54 2011