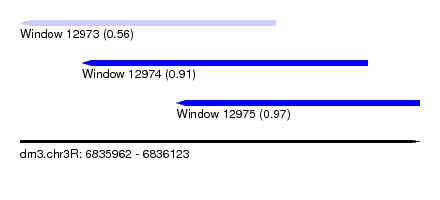
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,835,962 – 6,836,123 |
| Length | 161 |
| Max. P | 0.965333 |

| Location | 6,835,962 – 6,836,065 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Shannon entropy | 0.39718 |
| G+C content | 0.38942 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6835962 103 - 27905053 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAAGCGGCAGCAAUACGAUAGAAAACCAA------- .((((((......))))))....((((....)))).(((.((((((((((.(((..((..--((..-.........))..)))))))))))).))).)))......------- ( -21.30, z-score = -1.25, R) >droSim1.chr3R 12991052 103 + 27517382 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAAGCGCCAGCAAUACGAUAGAAAACCAA------- ....((((..((((((.((((..((((....)))).)))).(((((((((.((.......--))..-)))))))))))..((....)).......))))))))...------- ( -20.50, z-score = -1.41, R) >droSec1.super_0 5998601 103 + 21120651 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAAGCCA--GCAA-AACAACAAAGCAAGCGCCAGCAAUACGAUAGAAAACCAA------- ....((((..((((((.((((..((((....)))).)))).(((((((((.((.......--))..-)))))))))))..((....)).......))))))))...------- ( -20.50, z-score = -2.01, R) >droYak2.chr3R 10870327 105 - 28832112 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCAGCGCAA-AACAACAAAGCAGGCGGCAGCAAUACGAUAGAAAACCAU------- .((((((......))))))....((((....)))).(((.((((((((((.(((..(((.((....-.........)).))))))))))))).))).)))......------- ( -25.22, z-score = -1.73, R) >droEre2.scaffold_4770 2970299 103 + 17746568 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAGGCGGCAGCAGUACGAUAGAAAACCAA------- .((((((......))))))....((((....)))).(((...((((((((.(((..(((.--((..-.........)).))))))))))..))))..)))......------- ( -22.10, z-score = -0.81, R) >droAna3.scaffold_13340 11405845 79 + 23697760 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAGGC------------------------------- .((((((......))))))....((((....))))(((...(((((((((.((.......--))..-)))))))))..))).------------------------------- ( -16.60, z-score = -0.86, R) >droWil1.scaffold_181130 7566114 111 - 16660200 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAA-CAACAACAACAACAGCAACUAAAAAAACA-CGACACACCAGAUGUCCAUGAGGUGA .((((((......))))))....((((....))))(((((..((((((((....))-)))))).......................-.(((((....).))))..)))))... ( -20.70, z-score = -1.01, R) >droGri2.scaffold_14906 3705902 84 + 14172833 ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAA-CCA-AGCAGCCG-AGGAACAAGAACAACA-U------------------------- .((((((......))))))....((((....))))........(((((((......-((.-........-.))......)))))))-.------------------------- ( -15.00, z-score = -0.87, R) >consensus ACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA__GCAA_AACAACAAAGCAAGCGGCAGCAAUACGAUAGAAAACCAA_______ .((((((......))))))....((((....))))......(((((((((.((.........))...)))))))))..................................... (-13.40 = -13.85 + 0.45)
| Location | 6,835,987 – 6,836,102 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 88.63 |
| Shannon entropy | 0.22424 |
| G+C content | 0.37154 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -22.23 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6835987 115 - 27905053 ---CGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAAG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))(((...(((((((((.((.......--))..-)))))))))..))) ( -26.02, z-score = -2.09, R) >droSim1.chr3R 12991077 115 + 27517382 ---CGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAAG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))(((...(((((((((.((.......--))..-)))))))))..))) ( -26.02, z-score = -2.09, R) >droSec1.super_0 5998626 115 + 21120651 ---CGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAAGCCA--GCAA-AACAACAAAGCAAG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))(((...(((((((((.((.......--))..-)))))))))..))) ( -26.02, z-score = -2.51, R) >droYak2.chr3R 10870352 117 - 28832112 ---CGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCAGCGCAA-AACAACAAAGCAGG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))......(((((((((.(((......).))..-)))))))))..... ( -26.42, z-score = -1.51, R) >droEre2.scaffold_4770 2970324 115 + 17746568 ---CAGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAGG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))......(((((((((.((.......--))..-)))))))))..... ( -25.92, z-score = -1.98, R) >droAna3.scaffold_13340 11405846 115 + 23697760 ---CAGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA--GCAA-AACAACAAAGCAGG ---....(((((((((.............)))))))))...((((((......))))))....((((....))))......(((((((((.((.......--))..-)))))))))..... ( -25.92, z-score = -1.98, R) >dp4.chr2 2502112 110 + 30794189 CAGACCACCGCAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCAA------AACGGGGGG----- ....((((((((((((.............)))))))))...((((((......))))))...)))........((((((..((((..(((.....))))))------)..))))))----- ( -24.62, z-score = -0.35, R) >droPer1.super_6 3633436 110 - 6141320 CAGACCACCGCAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCAA------AACGGGGGG----- ....((((((((((((.............)))))))))...((((((......))))))...)))........((((((..((((..(((.....))))))------)..))))))----- ( -24.62, z-score = -0.35, R) >droWil1.scaffold_181130 7566154 106 - 16660200 -----GACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAA--CA--ACAACAACAACAG------ -----..(((((((((.............)))))))))...((((((......))))))....((((....)))).......((((((((....))--))--)))).........------ ( -24.02, z-score = -3.13, R) >droVir3.scaffold_13047 14162453 106 + 19223366 ---GCGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAA-CCA-------AGCAGCCGGG---- ---(((((((((((((.............)))))))))...((((((......))))))....((((....))))..)).....((((.....)))-)..-------.)).......---- ( -21.22, z-score = -0.51, R) >droGri2.scaffold_14906 3705917 106 + 14172833 ---ACGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAA-CCA-------AGCAGCCGAG---- ---....(((((((((.............)))))))))...((((((......))))))....((((....)))).........((((((......-...-------))))))....---- ( -21.02, z-score = -1.45, R) >consensus ___CCGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCAAUUUGUUGUUAGCCAGGCCA__GCAA_AACAACAAAGCA_G .......(((((((((.............)))))))))...((((((......))))))....((((....))))......(((((((((.................)))))))))..... (-22.23 = -21.48 + -0.76)
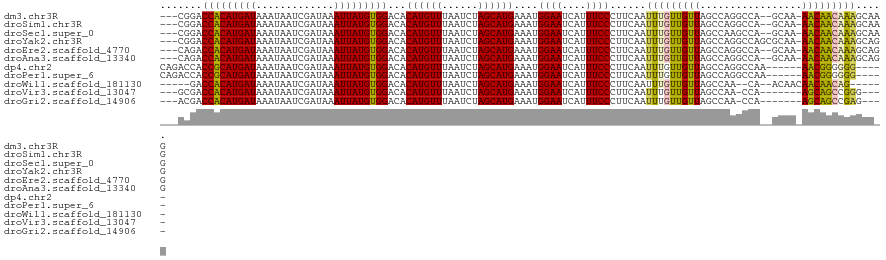
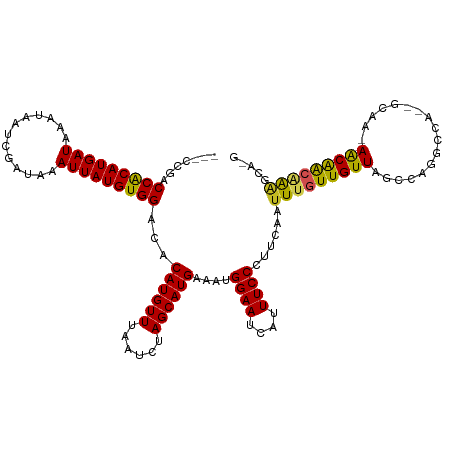
| Location | 6,836,025 – 6,836,123 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Shannon entropy | 0.33797 |
| G+C content | 0.37688 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -17.89 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6836025 98 - 27905053 -----------------CCACACAGAGCACA--CCCGUAGCGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA -----------------.........((((.--...)).))...(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -19.52, z-score = -1.22, R) >droSim1.chr3R 12991115 98 + 27517382 -----------------CCACACAGAGCACA--CCCGUAGCGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA -----------------.........((((.--...)).))...(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -19.52, z-score = -1.22, R) >droSec1.super_0 5998664 98 + 21120651 -----------------CCACACAGAGCACA--CCCGUAGCGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA -----------------.........((((.--...)).))...(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -19.52, z-score = -1.22, R) >droYak2.chr3R 10870392 100 - 28832112 ---------------CCACACAGAGAGCACA--CCCGUAGCGGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA ---------------.......(((.((((.--...)).))((((((((((((.............)))))))))...((((((......)))))).............))).))). ( -19.72, z-score = -1.17, R) >droEre2.scaffold_4770 2970362 98 + 17746568 -----------------CCACACAGAGCACA--CCCGUAGCAGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA -----------------.........((((.--...)).))...(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -19.52, z-score = -1.76, R) >droAna3.scaffold_13340 11405884 98 + 23697760 -----------------CCACACAGAGCACA--CCCGUAGCAGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA -----------------.........((((.--...)).))...(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -19.52, z-score = -1.76, R) >dp4.chr2 2502142 116 + 30794189 AGAGCGAACCUAAGCGUACACACAGACAACA-GACAACAGACCACCGCAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA .((((........))................-.........((((((((((((.............)))))))))...((((((......))))))...)))..))........... ( -19.42, z-score = -1.01, R) >droPer1.super_6 3633466 116 - 6141320 AGAGCGAACCUAAGCGUACACACAGACAACA-GACAACAGACCACCGCAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA .((((........))................-.........((((((((((((.............)))))))))...((((((......))))))...)))..))........... ( -19.42, z-score = -1.01, R) >droWil1.scaffold_181130 7566185 78 - 16660200 ---------------------------------------CCAGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA ---------------------------------------.....(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -17.82, z-score = -2.43, R) >droVir3.scaffold_13047 14162482 91 + 19223366 --------------------------CUACAAACAAACAGGCGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA --------------------------..................(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -17.82, z-score = -2.01, R) >droMoj3.scaffold_6540 21366286 91 - 34148556 --------------------------CGACAAACAAACGGGCGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA --------------------------............(((...(((((((((.............)))))))))...((((((......))))))(((((....)))))))).... ( -19.12, z-score = -1.82, R) >droGri2.scaffold_14906 3705946 91 + 14172833 --------------------------CCACAAUCAAACAGACGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA --------------------------..................(((((((((.............)))))))))...((((((......))))))....((((....))))..... ( -17.82, z-score = -1.95, R) >consensus _________________CCACACAGAGCACA__CCCGUAGCCGACCACAUGAUAAAUAAUCGAUAAAUUAUGUGGACACAUGUUUAAUCUAGCAUGAAAUGGAAUCAUUUCCCUUCA ............................................(((((((((.............)))))))))...((((((......))))))....((((....))))..... (-17.89 = -17.75 + -0.14)
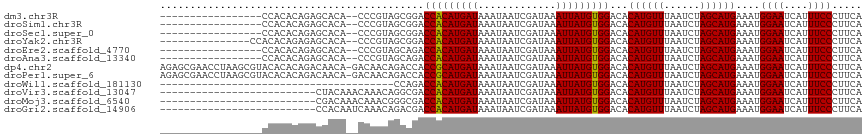
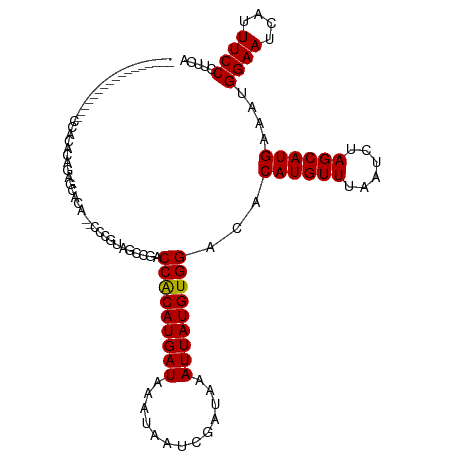
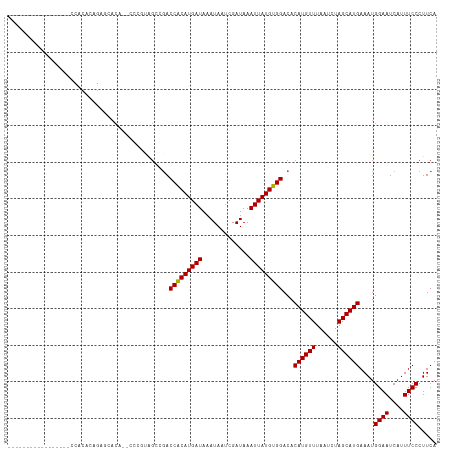
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:51 2011