| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,807,635 – 6,807,739 |
| Length | 104 |
| Max. P | 0.868258 |

| Location | 6,807,635 – 6,807,739 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.60183 |
| G+C content | 0.40073 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
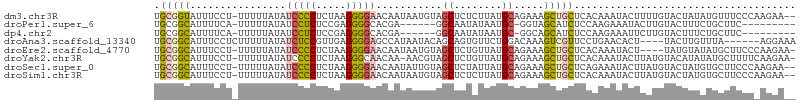
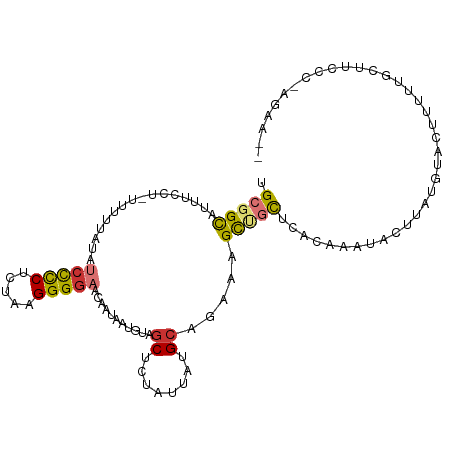
>dm3.chr3R 6807635 104 - 27905053 UGCGGUAUUUCCU-UUUUUAUAUCCCCUCUAAGGGGAACAAUAAUGUAGCUCUCUUAUGCAGAAAGCUGCUCACAAAUACUUUUGUACUAUAUGUUUCCCAAGAA-- ...((.....)).-..................((((((((.....((((((.(((.....))).))))))..(((((....)))))......)))))))).....-- ( -24.90, z-score = -2.48, R) >droPer1.super_6 3592868 90 - 6141320 UGCGGCAUUUUCA-UUUUUAUAUCCUCUCCGAGGGGCACGA------GGCAAUAUAAUGC-GGUAGCAUCUCCAAGAAAUACUUGUACUUUCUGCUUC--------- .((((........-........(((((...)))))..((((------((((......)))-((........))........))))).....))))...--------- ( -16.20, z-score = 0.65, R) >dp4.chr2 28257090 90 - 30794189 UGCGGCAUUUUCA-UUUUUAUAUCCUCUCCGAGGGGCACGA------GGCAAUAUAAUGC-GGCAGCAUCUCCAAGAAAUUCUUGUACUUUCUGCUUC--------- (((.(((((....-........(((((...)))))((....------.)).....)))))-.)))(((....((((.....)))).......)))...--------- ( -18.70, z-score = 0.05, R) >droAna3.scaffold_13340 11378492 97 + 23697760 UGCGGCAUUUCCUCUUUUUAUAUCUCCGUUGAGGGGAGCCAUAAUACAGCAGUGUUCUGGACAAAGUCGUUCCUGAACACU----UACUUGUUUA------AGGAAA .(((((..((((((((..............))))))))(((.(((((....))))).))).....)))))(((((((((..----....))))).------)))).. ( -26.64, z-score = -1.67, R) >droEre2.scaffold_4770 2941825 101 + 17746568 UGCGGCAUUUCCU-UUUUUAUAUCCCCUCUAAGGGGAACAAUAAUGUAGCUCUGUUAUGCAGAAAGCUGCUCACAAAUACU----UAUGUAUAUGCUUCCCAAGAA- ...(((((.....-........(((((.....)))))........(((((((((.....))))..)))))......((((.----...))))))))).........- ( -24.00, z-score = -1.59, R) >droYak2.chr3R 10841149 104 - 28832112 UGCGGCAUUUCCU-UUUUUAUAUCCCCUCUAAGGGCAACAA-AACGUAGCUCUGUUAUGCAGAAAGCUGCUCACAAAUACUUAUGUACAUAUAUGCUUUUCAAGAA- .............-............((..(((((((....-...(((((((((.....))))..))))).......(((....)))......)))))))..))..- ( -17.70, z-score = 0.08, R) >droSec1.super_0 5969667 104 + 21120651 UGCGGCAUUUCCU-UUUUUAUAUCCCCUCUAAGGGGAACAAUAUUGUAGCUCUAUUAUGCAGAAAGCUGCUCAGAAAUACUUAUGUACUAUGUGCUUCCCAAGAA-- .(((((.((((..-........(((((.....)))))...........((........)).)))))))))..............((((...))))..........-- ( -22.30, z-score = -0.57, R) >droSim1.chr3R 12961321 104 + 27517382 UGCGGCAUUUCCU-UUUUUAUAUCCCCUCUAAGGGGAACAAUAAUGUAGCUCUCUUAUGCAGAAAGCUGCUCACAAAUACUUAUGUACUAUGUGCUUCCCAAGAA-- .(((((.((((..-........(((((.....)))))...........((........)).))))))))).((((..(((....)))...))))...........-- ( -24.20, z-score = -1.61, R) >consensus UGCGGCAUUUCCU_UUUUUAUAUCCCCUCUAAGGGGAACAAUAAUGUAGCUCUAUUAUGCAGAAAGCUGCUCACAAAUACUUAUGUACUUUUUGCUUCCC_AGAA__ .(((((................(((((.....)))))...........((........)).....)))))..................................... (-12.91 = -13.21 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:46 2011