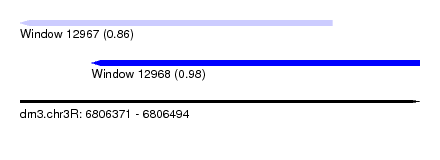
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,806,371 – 6,806,494 |
| Length | 123 |
| Max. P | 0.980440 |

| Location | 6,806,371 – 6,806,467 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 61.09 |
| Shannon entropy | 0.76720 |
| G+C content | 0.40246 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -6.85 |
| Energy contribution | -6.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862699 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 6806371 96 - 27905053 AUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC---------UUGGGUUGGCGCCCCCACUCGCUCCACUUGCC-----------AUUUUUGUCUAUUCAU (((((((((............)))))))))((((((((((........---------..))))))))))........((.......)).-----------................ ( -17.40, z-score = -0.81, R) >droSim1.chr3R 12960060 95 + 27517382 AUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC---------UUGGGUUCGCG-CCCCACUCGCUCCACUUGCC-----------AUUUUUGUCUAUUCAU (((((((((............)))))))))((((.......((((...---------.))))...)))-).......((.......)).-----------................ ( -13.40, z-score = -0.03, R) >droSec1.super_0 5968408 95 + 21120651 AUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC---------UUGGGUUCGCG-CCCCACUCGCUUCCCUUGCC-----------AUUUUUGUCUAUUCAU (((((((((............)))))))))((((.......((((...---------.))))...)))-).......((.......)).-----------................ ( -13.40, z-score = -0.16, R) >droYak2.chr3R 10839892 105 - 28832112 AUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC---------UUGGAUUCGCA-CCGCACUUGCCCCAUUCGCUGC-CACUUGCCAUUUUUGUCUAUUCAU ((((((((((..........))))))))))..................---------.(((((..(((-..((....)).......((...-.....))......)))..))))). ( -14.30, z-score = 0.47, R) >droEre2.scaffold_4770 2940592 94 + 17746568 AUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC---------UUGGGCUCGCG-CCCCACUCGCCCCACUUGCC------------UUUUUGUCUAUUCAU ((((((((((..........))))))))))..((.........((...---------..((((....)-)))....))........)).------------............... ( -15.73, z-score = -0.73, R) >droAna3.scaffold_13340 11377380 115 + 23697760 AUGAUUUUCAAUUUUAUGCCUGAAAGUCAUGCGCUAAUUUUACCAUUUUCC-UCUGUCCAAGUUCUAAGUUCCCCAAGCUUUUUCUUUCUCGCCCGGGACGUUUCUGUCUAUUCAU ((((((((((..........)))))))))).....................-...(((((((....(((((.....)))))...)))...(....)))))................ ( -14.20, z-score = 0.05, R) >dp4.chr2 28254565 89 - 30794189 AUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUCCUCCAC---------CGGGGU------UUUUUCUUUCGGUGUUUC-U-----------UUUUCUGUCUAUUCAU ((((....((......((((((.....))))))............(((---------((..(.------......)..)))))....-.-----------.....)).....)))) ( -18.10, z-score = -2.81, R) >droPer1.super_6 3590510 90 - 6141320 AUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUCCUCCAC---------CGGGGU------UUUUUCUUUCGGUGUUUCUU-----------UUUUCUGUCUAUUCAU ((((....((......((((((.....))))))............(((---------((..(.------......)..)))))......-----------.....)).....)))) ( -18.10, z-score = -2.85, R) >droWil1.scaffold_181130 7536364 81 - 16660200 AUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUGUUGUCU---------------------GCCUGCU-GCUUCUAUUGCC-------------UUUUGCCUAUUCAU ..(((...(((..(((((((((.....)))))).)))..)))..))).---------------------((.....-((.......)).-------------....))........ ( -15.20, z-score = -1.43, R) >droVir3.scaffold_13047 14126168 72 + 19223366 AUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUCUUGUUUGCCAUUU--GCAAUUCGCCUAUUCAU------------------------------------------ ((((............((((((.....)))))).............((((.....--)))).........))))------------------------------------------ ( -10.80, z-score = -0.16, R) >droMoj3.scaffold_6540 21329045 72 - 34148556 AUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUCUUGUUUGCUAUUU--GCAAUUCGCCUAUUCAU------------------------------------------ ((((............((((((.....)))))).............((((.....--)))).........))))------------------------------------------ ( -12.00, z-score = -0.68, R) >droGri2.scaffold_14906 3669404 82 + 14172833 AUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUGUUGUUUGCCAUUUCGCCAGUUCGCCAGUUCGCCUAUUCAU---------------------------------- ((((....(((..(((((((((.....)))))).)))..)))......((...((.((......)).))...))....))))---------------------------------- ( -12.30, z-score = 0.41, R) >consensus AUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCC_________UUGGGUUCGCG_UCCCACUCGCUCCACUUGCC____________UUUUUGUCUAUUCAU (((((((((............)))))))))...................................................................................... ( -6.85 = -6.85 + 0.00)


| Location | 6,806,393 – 6,806,494 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.34 |
| Shannon entropy | 0.66906 |
| G+C content | 0.47296 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -12.26 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6806393 101 - 27905053 ----------CCACUCGAGGCGUCAGGUGUUGGCGCAAUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUUGGGUUGGCGCCCCCACUCGCUCCA----- ----------..((((((((((..(((.(((((((((.((((((((............)))))))))))))))))..))))).))))))))(((.((........)).)))----- ( -36.00, z-score = -2.87, R) >droSim1.chr3R 12960082 100 + 27517382 ----------CCACUCGAGGCGUCAGGUGUUGGCGCAAUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUUGGGUUCGCG-CCCCACUCGCUCCA----- ----------..((((((((((..(((.(((((((((.((((((((............)))))))))))))))))..))))).))))))))..(((-.......)))....----- ( -34.10, z-score = -2.81, R) >droSec1.super_0 5968430 100 + 21120651 ----------CCACUCGAGGCGUCAGGUGUUGGCGCAAUGAUUUUCCAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUUGGGUUCGCG-CCCCACUCGCUUCC----- ----------..((((((((((..(((.(((((((((.((((((((............)))))))))))))))))..))))).))))))))..(((-.......)))....----- ( -34.10, z-score = -2.95, R) >droYak2.chr3R 10839924 100 - 28832112 ----------CCACUUGAGGCGUCAGGUGUUGCCGCAAUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUUGGAUUCGCA-CCGCACUUGCCCCA----- ----------........((.(.((((((((((((((.(((((((((..........)))))))))))))...........(((....)))..)))-..))))))).))).----- ( -27.80, z-score = -1.67, R) >droEre2.scaffold_4770 2940613 100 + 17746568 ----------ACACUCGAGGCGUCAGGUGUUGCCGCAAUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUUGGGCUCGCG-CCCCACUCGCCCCA----- ----------......(.((((...(((((((.((((.(((((((((..........))))))))))))).))).............((((....)-))))))))))).).----- ( -29.50, z-score = -1.60, R) >droAna3.scaffold_13340 11377416 93 + 23697760 -----------------------CAGGUGUUGCUGCAAUGAUUUUCAAUUUUAUGCCUGAAAGUCAUGCGCUAAUUUUACCAUUUUCCUCUGUCCAAGUUCUAAGUUCCCCAAGCU -----------------------..((((..((.(((.(((((((((..........))))))))))))))......))))................................... ( -16.40, z-score = -0.89, R) >dp4.chr2 28254581 103 - 30794189 -------ACGCCACUCGCGUUGUCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUCCUCCACCGGGGUUUUUUCUUUCGGUGUUUCU------ -------((((.....))))....(((.((((.((((.(((((((((..........))))))))))))).))))...))).(((((..(.......)..))))).....------ ( -27.40, z-score = -1.82, R) >droPer1.super_6 3590527 103 - 6141320 -------ACGCCACUCGCGUUGUCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUCCUCCACCGGGGUUUUUUCUUUCGGUGUUUCU------ -------((((.....))))....(((.((((.((((.(((((((((..........))))))))))))).))))...))).(((((..(.......)..))))).....------ ( -27.40, z-score = -1.82, R) >droWil1.scaffold_181130 7536384 97 - 16660200 AUGAAGGCGC-CACUUGAGCUGGCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCAUGAAAGUCAUGCGCUAAUUUUGUUGUCUGCCUGCUGCUUCU------------------ ..(((((((.-((((((......)))))).))).(((..(((...(((..(((((((((.....)))))).)))..)))..)))....)))..)))).------------------ ( -29.90, z-score = -1.05, R) >droVir3.scaffold_13047 14126184 90 + 19223366 ---AAGCUGCUUGCCUCAAGUGGCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUCUUGUUUGCCAUUUG----------------------- ---.(((.(((((((......)))))))...)))(((.(((((((((..........))))))))))))((.(((......))).))......----------------------- ( -26.10, z-score = -1.50, R) >droMoj3.scaffold_6540 21329061 91 - 34148556 --AGAGCCACUUGCUUGAAGUGGCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUCUUGUUUGCUAUUUG----------------------- --..(((((((((((......))))))))..)))(((.(((((((((..........))))))))))))((.(((......))).))......----------------------- ( -25.90, z-score = -1.42, R) >droGri2.scaffold_14906 3669425 96 + 14172833 --AAAGCUGCUUGCCUCAAGUGGCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCGUGAAAGUCAUGCGCUAAUUUUGUUGUUUGCCAUUUCGCCAG------------------ --...((..((((...))))..)).((((.....((((.......(((..(((((((((.....)))))).)))..)))....)))).....))))..------------------ ( -29.60, z-score = -1.68, R) >consensus __________CCACUCGAGGUGUCAGGUGUUGCUGCAAUGAUUUUCAAUUUUACGCCUGAAAGUCAUGCGCUAAUUUUCUCGUCCUCGGGUUCGCG_CC_CACU_GCUCC______ ..................((........((((.((((.((((((((............)))))))))))).))))........))............................... (-12.26 = -11.58 + -0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:45 2011