
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,796,928 – 6,797,045 |
| Length | 117 |
| Max. P | 0.999032 |

| Location | 6,796,928 – 6,797,045 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.54516 |
| G+C content | 0.36468 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -11.17 |
| Energy contribution | -13.26 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6796928 117 - 27905053 AUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAAA ...((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))..............((........))............ ( -26.20, z-score = -5.22, R) >droSim1.chr3R 12950486 116 + 27517382 AUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAA- ...((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))..............((........))...........- ( -26.20, z-score = -5.30, R) >droSec1.super_0 5959256 116 + 21120651 AUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACCCAUCACACCGCUUACAAAAAAA- ...((((((((((......)))))))))).......((((((((.(((....((((...))))....))).)))))))).....................................- ( -25.40, z-score = -5.26, R) >droYak2.chr3R 10830266 116 - 28832112 CUUAAUUAAUCAGAAUUUCCCGAUUGAUUUCGUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAA- ...((((((((.(......).)))))))).......((((((((.(((....((((...))))....))).))))))))..............((........))...........- ( -21.60, z-score = -3.19, R) >droEre2.scaffold_4770 2931004 116 + 17746568 AUUAAUUAAUCAGAAUUUUCUGAUUGAUUUCGUCAAGGUUUUGGCUAAAAAAUGGCACCGCUACAACUGAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAA- ...(((((((((((....))))))))))).......(((((((((((.....))))...........((......)).......)))))))..((........))...........- ( -17.80, z-score = -1.46, R) >droMoj3.scaffold_6540 21317289 96 - 34148556 AUUCAUUAAUGAAAUUGUCUUUAUGCUUUUUCGAA--AUAUGCAUCAAAAAG-GGCAUAAACUGAA---AAACUAUCGUACUAU-GUAUUUACGAUUGGCGUC-------------- .((((....)))).........(((((((((((..--.(((((.((.....)-))))))...))))---)))(.((((((....-.....)))))).))))).-------------- ( -14.90, z-score = -0.52, R) >droGri2.scaffold_14906 3657649 97 + 14172833 AUUCAUUAAUAGGAUUUUCUUUAUGCUUUUCGAAAUGCUGUGCGAAAAAAAG-GGCACAAG-GGAG---AAACUACCGUACUAU-GUACUUACGACUGUCCGC-------------- ...........(((((((((((.(((((((.....(((...))).....)))-)))).)))-))))---).......((((...-)))).........)))..-------------- ( -16.80, z-score = 0.39, R) >consensus AUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAA_ ...((((((((((......)))))))))).......((((((((........((((...))))........))))))))...................................... (-11.17 = -13.26 + 2.09)

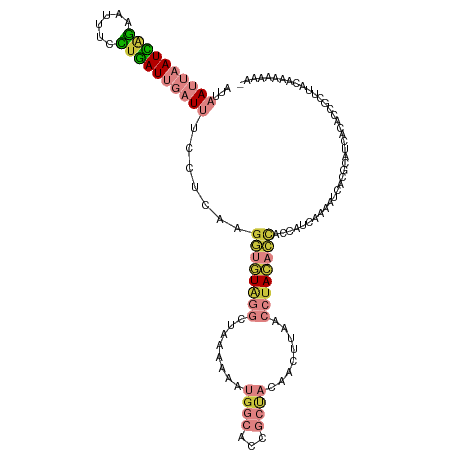
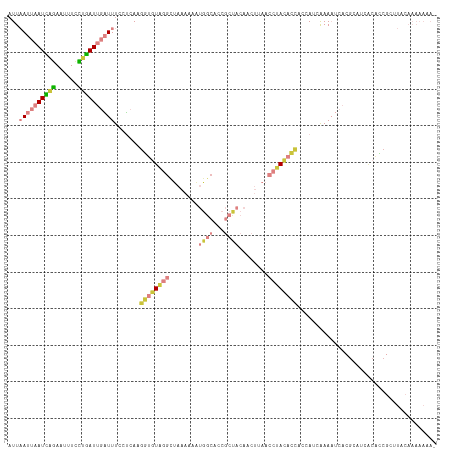
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:44 2011