| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,779,547 – 6,779,640 |
| Length | 93 |
| Max. P | 0.877231 |

| Location | 6,779,547 – 6,779,640 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.97 |
| Shannon entropy | 0.08751 |
| G+C content | 0.52845 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
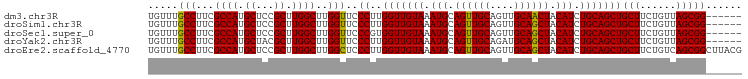
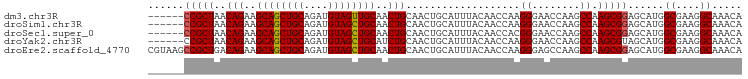
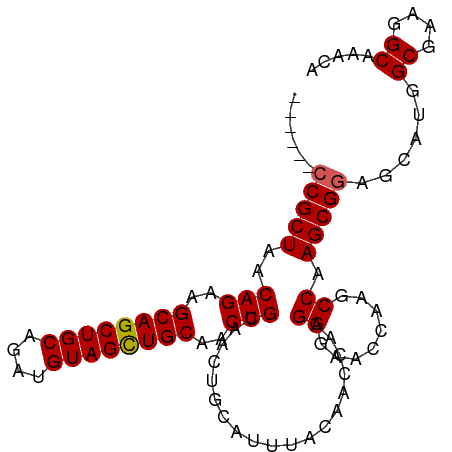
>dm3.chr3R 6779547 93 + 27905053 UGUUUGCCUUCGCCAUGCUCCGCUUGGCUUGGUUCCCUUGGUUGUAAAUGCAGUUGCAGUUGCAACUACAUCUGCAGCUGCUUCUGUUAGCGG------ .....(((...((((.((...)).))))..)))..((..(((((((.(((.((((((....)))))).))).)))))))(((......)))))------ ( -31.00, z-score = -1.50, R) >droSim1.chr3R 12937574 93 - 27517382 UGUUUGCCUUCGCCAUGCUCCGCUUGGCUUGGUUCCCUUGGUUGUAAAUGCAGUUGCAGUUGCAGCUACAUCUGCAGCUGCUUCUGUUAGCGG------ .((((((..((((((.((...)).))))..((...))..))..))))))((((..((((((((((......))))))))))..))))......------ ( -31.80, z-score = -1.12, R) >droSec1.super_0 5947463 93 - 21120651 UGUUUGCCUUCGCCAUGCUCCGCUUGGCUUGGUUCCCGUGGUUGUAAAUGCAGUUGCAGUUGCAGCUACAUCUGCAGCUGCUUCUGUUAGCGG------ .....(((...((((.((...)).))))..)))..((((..........((((..((((((((((......))))))))))..))))..))))------ ( -34.20, z-score = -1.73, R) >droYak2.chr3R 10817769 93 + 28832112 UGUUUGCCUUCGCCAUGCUACGCUUGGCUUGGUUCCCUUGGUUGUAAAUGCAGUUGCAGAUGCAGCUACAUCUGCAGCUGCUUCUGUUAGCGG------ .....(((...((((.((...)).))))..))).......((((.....(((((((((((((......)))))))))))))......))))..------ ( -34.50, z-score = -1.80, R) >droEre2.scaffold_4770 2918969 99 - 17746568 UGUUUGCCUUCGCCAUGCUCCGCUUGGCUUGGCUCCCUUGGUUGUAAAUGCAGUUGCAGUUGCAGCUACAUCUGCAGCUGCUUCUGUCAGCGGCUUACG .((..(((...((((.(((......))).)))).......((((.....((((..((((((((((......))))))))))..)))))))))))..)). ( -40.50, z-score = -2.88, R) >consensus UGUUUGCCUUCGCCAUGCUCCGCUUGGCUUGGUUCCCUUGGUUGUAAAUGCAGUUGCAGUUGCAGCUACAUCUGCAGCUGCUUCUGUUAGCGG______ .....(((...((((.((...)).))))..)))..((..(((((((.(((.((((((....)))))).))).)))))))(((......)))))...... (-31.82 = -31.50 + -0.32)
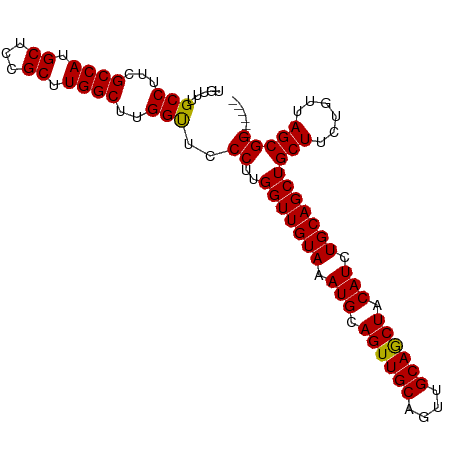
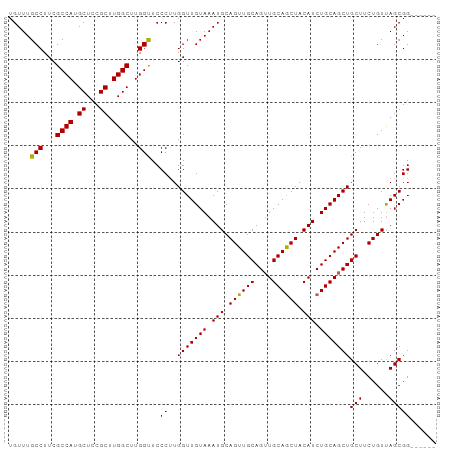
| Location | 6,779,547 – 6,779,640 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Shannon entropy | 0.08751 |
| G+C content | 0.52845 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6779547 93 - 27905053 ------CCGCUAACAGAAGCAGCUGCAGAUGUAGUUGCAACUGCAACUGCAUUUACAACCAAGGGAACCAAGCCAAGCGGAGCAUGGCGAAGGCAAACA ------(((((.......((....))((((((((((((....))))))))))))........((........)).)))))......((....))..... ( -29.90, z-score = -1.78, R) >droSim1.chr3R 12937574 93 + 27517382 ------CCGCUAACAGAAGCAGCUGCAGAUGUAGCUGCAACUGCAACUGCAUUUACAACCAAGGGAACCAAGCCAAGCGGAGCAUGGCGAAGGCAAACA ------(((((..(((..((((((((....))))))))..)))...................((........)).)))))......((....))..... ( -30.20, z-score = -1.59, R) >droSec1.super_0 5947463 93 + 21120651 ------CCGCUAACAGAAGCAGCUGCAGAUGUAGCUGCAACUGCAACUGCAUUUACAACCACGGGAACCAAGCCAAGCGGAGCAUGGCGAAGGCAAACA ------(((((..(((..((((((((....))))))))..)))...................((........)).)))))......((....))..... ( -30.30, z-score = -1.65, R) >droYak2.chr3R 10817769 93 - 28832112 ------CCGCUAACAGAAGCAGCUGCAGAUGUAGCUGCAUCUGCAACUGCAUUUACAACCAAGGGAACCAAGCCAAGCGUAGCAUGGCGAAGGCAAACA ------..(((.......((((.(((((((((....))))))))).)))).....................((((.((...)).))))...)))..... ( -31.80, z-score = -2.03, R) >droEre2.scaffold_4770 2918969 99 + 17746568 CGUAAGCCGCUGACAGAAGCAGCUGCAGAUGUAGCUGCAACUGCAACUGCAUUUACAACCAAGGGAGCCAAGCCAAGCGGAGCAUGGCGAAGGCAAACA .((((((.(.((.(((..((((((((....))))))))..))))).).))..))))..........(((..((((.((...)).))))...)))..... ( -36.00, z-score = -1.85, R) >consensus ______CCGCUAACAGAAGCAGCUGCAGAUGUAGCUGCAACUGCAACUGCAUUUACAACCAAGGGAACCAAGCCAAGCGGAGCAUGGCGAAGGCAAACA ......(((((..(((..((((((((....))))))))..)))...................((........)).)))))......((....))..... (-28.46 = -28.50 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:41 2011