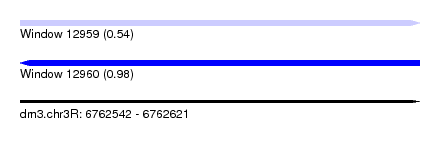
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,762,542 – 6,762,621 |
| Length | 79 |
| Max. P | 0.976076 |

| Location | 6,762,542 – 6,762,621 |
|---|---|
| Length | 79 |
| Sequences | 4 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.41602 |
| G+C content | 0.59456 |
| Mean single sequence MFE | -18.98 |
| Consensus MFE | -9.54 |
| Energy contribution | -10.85 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537153 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
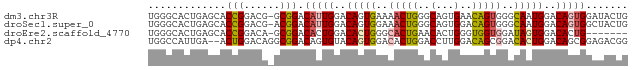
>dm3.chr3R 6762542 79 + 27905053 CAGUAUCCACUGUCCAUUGCCCACUGUUCACUGCCCAGUUUUCACUGUCCAAUGUCCGC-CGUCCGGUGCUCAGUGCCCA .......(((((..(((((..(..((..((.((..((((....))))..)).))..)).-.)..)))))..))))).... ( -17.80, z-score = -1.77, R) >droSec1.super_0 5930549 79 - 21120651 CAGUAGCCACUGUCCAUUGCCCACUGUCCACUGCCCAGUUUCCACUGUCCAAUGUCCGU-CGUCCGGUGCUCAGUGCCCA .......(((((..(((((..(..((..((.((..((((....))))..)).))..)).-.)..)))))..))))).... ( -18.10, z-score = -1.41, R) >droEre2.scaffold_4770 2901427 72 - 17746568 -------CAGUGUCCACUAUCCACCACCCAGUGUUCAGUGCCCAGUGUCCAGUGUCCGC-UGUCCGGUGCUCAGUGCCCA -------...((..((((...((((...(((((..((.((.........)).))..)))-))...))))...))))..)) ( -18.80, z-score = -1.53, R) >dp4.chr2 7173564 78 + 30794189 CCGUCUCCGCUGUCCAGUGUCCGCUGUCCAAGGUCCAGUGUCCACUGUACACUGUCCGCCUGUCCAGU--UCAAUGGCCA ........(((((.(((((..(((((.(....)..)))))..))))).))((((..(....)..))))--.....))).. ( -21.20, z-score = -2.21, R) >consensus CAGUAUCCACUGUCCAUUGCCCACUGUCCACUGCCCAGUGUCCACUGUCCAAUGUCCGC_CGUCCGGUGCUCAGUGCCCA .......(((((..(((((..(..((..((.((..((((....))))..)).))..))...)..)))))..))))).... ( -9.54 = -10.85 + 1.31)


| Location | 6,762,542 – 6,762,621 |
|---|---|
| Length | 79 |
| Sequences | 4 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.41602 |
| G+C content | 0.59456 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976076 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
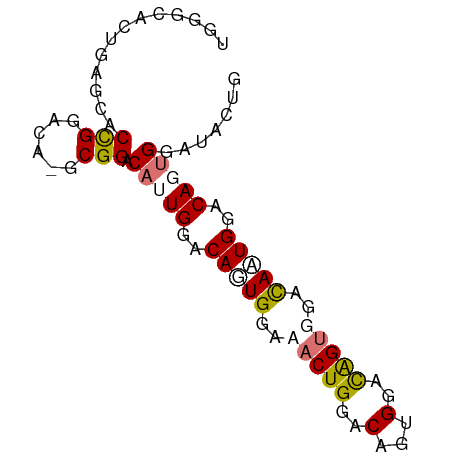
>dm3.chr3R 6762542 79 - 27905053 UGGGCACUGAGCACCGGACG-GCGGACAUUGGACAGUGAAAACUGGGCAGUGAACAGUGGGCAAUGGACAGUGGAUACUG ((..(((((....(((....-.))).(((((..((((....))))..)))))..)))))..)).....((((....)))) ( -24.10, z-score = -1.96, R) >droSec1.super_0 5930549 79 + 21120651 UGGGCACUGAGCACCGGACG-ACGGACAUUGGACAGUGGAAACUGGGCAGUGGACAGUGGGCAAUGGACAGUGGCUACUG (((.(((((....(((..(.-(((..(((((..((((....))))..)))))..).)).)....))).))))).)))... ( -28.80, z-score = -3.04, R) >droEre2.scaffold_4770 2901427 72 + 17746568 UGGGCACUGAGCACCGGACA-GCGGACACUGGACACUGGGCACUGAACACUGGGUGGUGGAUAGUGGACACUG------- .((((.....)).))...((-(.(..(((((..(((....((((.(....).)))))))..)))))..).)))------- ( -22.40, z-score = -1.04, R) >dp4.chr2 7173564 78 - 30794189 UGGCCAUUGA--ACUGGACAGGCGGACAGUGUACAGUGGACACUGGACCUUGGACAGCGGACACUGGACAGCGGAGACGG ((.(((.((.--.(((..((((.(..((((((.......))))))..)))))..)))....)).))).)).((....)). ( -27.60, z-score = -2.81, R) >consensus UGGGCACUGAGCACCGGACA_GCGGACAUUGGACAGUGGAAACUGGACAGUGGACAGUGGACAAUGGACAGUGGAUACUG ....(((((....(((......))).(((((..((((....))))..)))))..)))))..................... (-16.12 = -16.12 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:39 2011