| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,754,269 – 6,754,363 |
| Length | 94 |
| Max. P | 0.544133 |

| Location | 6,754,269 – 6,754,363 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.48664 |
| G+C content | 0.61812 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
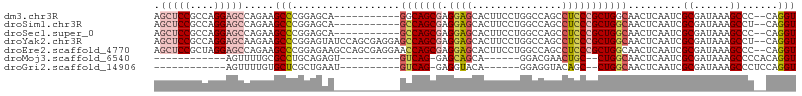
>dm3.chr3R 6754269 94 + 27905053 AGCUCCGCCAGGAGCCAGAAGCCCGGAGCA-----------GGCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCC--CAGGU .(((((....))))).....(((.((.((.-----------....((((((((...))))..((((((.....)))))).......))))......)).)--).))) ( -35.40, z-score = -0.35, R) >droSim1.chr3R 12912692 94 - 27517382 AGCUCCGCCAGGAGCCAGAAGCCCGGAGCA-----------GCCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCU--CAGGU .(((((....))))).....(((.((((..-----------(((((((.(((((............).)))))))))))..)))....((......))..--).))) ( -35.60, z-score = -0.66, R) >droSec1.super_0 5922198 94 - 21120651 AGCUCCGCCAGGAGCCAGAAGCCCGGAGCA-----------GCCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCC--CAGGU .(((((....))))).....(((.((.((.-----------(((((((.(((((............).)))))))))))...((......))....)).)--).))) ( -36.70, z-score = -1.21, R) >droYak2.chr3R 10791415 105 + 28832112 AGCUCCGCCAGGAGCAAGAAGCCCGGAGUAUCCAGCGAGGAGCCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCU--CAGGU .(((((....))))).....(((.(((((..((((((.(((((..((.(((((...))))).))..).))))))))))..))))....((......))..--).))) ( -44.60, z-score = -2.27, R) >droEre2.scaffold_4770 2893026 105 - 17746568 AGCUCCGCUAGGAGCCAGAAGCCCGGAGAAGCCAGCGAGGAACCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCC--CAGGU .(((((....))))).....(((.((((..((((((((((..((((.((((....))))))))....))))..))))))..)))....((......))..--).))) ( -43.00, z-score = -1.93, R) >droMoj3.scaffold_6540 21272039 76 + 34148556 ------------AGUUUUGCGCCUGCAGAGU----------GUCAG-GAGCAGCA------GGACGAACUGC--CUGGCAACUCAAUCGCGAUAAAGCCCCACAGGU ------------........(((((..((((----------(((((-.....(((------(......))))--))))).))))....((......))....))))) ( -23.10, z-score = -0.21, R) >droGri2.scaffold_14906 3617745 76 - 14172833 ------------AGUUUUGUGCUCGCUGAAU----------GUCAG-GAGGUACA------GGAGGUACAGC--CUGGCAACUCAAUCGCGAUAAAGCCCUCCAGGU ------------.((((((...(((((((.(----------(((((-(..((((.------....))))..)--))))))..)))...))))))))))((....)). ( -24.10, z-score = -0.99, R) >consensus AGCUCCGCCAGGAGCCAGAAGCCCGGAGAA___________GCCAGCGAGGAGCACUUCCUGGCCAGCCUCCCGCUGGCAACUCAAUCGCGAUAAAGCCC__CAGGU .(((((....))))).....(((..................(((((((.((((...............))))))))))).........((......))......))) (-19.66 = -20.86 + 1.20)

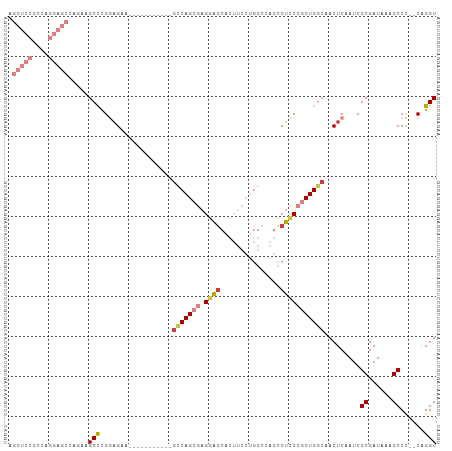
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:37 2011