
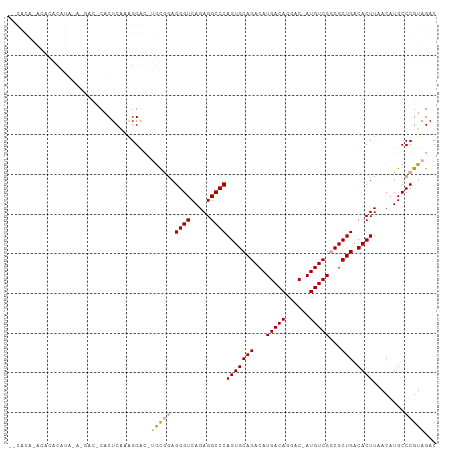
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,749,199 – 6,749,324 |
| Length | 125 |
| Max. P | 0.888981 |

| Location | 6,749,199 – 6,749,301 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.35164 |
| G+C content | 0.55438 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -22.51 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6749199 102 + 27905053 --CACACACACACAUAUAAG---CACUCAAAGGACCUGCGGAGGGUCAGAGGCCCAGUGCAGACAUGACAGGAC-AUGUCGGCGCUGACACUUAACAUGCCCGUAGAC --..................---............((((((.((((.....))))(((((.((((((......)-))))).)))))..............)))))).. ( -31.70, z-score = -2.10, R) >droYak2.chr3R 10786210 107 + 28832112 UACACAAACACACAUAAAGGACACACUCACAGGACAUGCGGAGGGUCAGAGGCCCAGUGCAGACAUGACAGGAC-AUGUCGGCGCUGACACUUAACAUGCCCGUAGAC .........................((.((.((.((((..((((((.....)))((((((.((((((......)-))))).))))))...)))..)))))).)))).. ( -31.80, z-score = -2.62, R) >droAna3.scaffold_13340 11326716 86 - 23697760 -------------------CACUCACUCGUGGGG---AUAUAGGGUCAGAGGCCCAGUGCAGACAUGACAGGCCCAUGUCGCCGCUGACACUUAACAUGCCCGUAGAC -------------------(((......)))(((---...(((((((((.(((........((((((.......))))))))).))))).)))).....)))...... ( -26.90, z-score = -0.26, R) >consensus __CACA_ACACACAUA_A_GAC_CACUCAAAGGAC_UGCGGAGGGUCAGAGGCCCAGUGCAGACAUGACAGGAC_AUGUCGGCGCUGACACUUAACAUGCCCGUAGAC ....................................(((((.((((.....))))(((((((...(((((......)))))...))).))))........)))))... (-22.51 = -23.07 + 0.56)


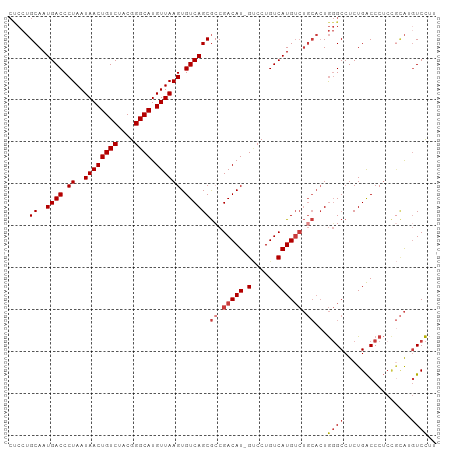
| Location | 6,749,223 – 6,749,324 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.15 |
| Shannon entropy | 0.22089 |
| G+C content | 0.54810 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -20.25 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6749223 101 - 27905053 CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU-GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAGGUCCUU ..(((((...(((.((..((((((((....)))).)))))).)))((.((.(((((-(......)))))).)).))(((.......)))...)))))..... ( -35.20, z-score = -3.03, R) >droSim1.chr3R 12907616 101 + 27517382 CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU-GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAGGUCCUU ..(((((...(((.((..((((((((....)))).)))))).)))((.((.(((((-(......)))))).)).))(((.......)))...)))))..... ( -35.20, z-score = -3.03, R) >droSec1.super_0 5917194 101 + 21120651 CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU-GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAGGUCCUU ..(((((...(((.((..((((((((....)))).)))))).)))((.((.(((((-(......)))))).)).))(((.......)))...)))))..... ( -35.20, z-score = -3.03, R) >droYak2.chr3R 10786239 101 - 28832112 CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU-GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAUGUCCUG ..........(((..........)))...((((((((..((.(((((.((((((((-(......)))))).......)))..))))).))..)))))))).. ( -30.71, z-score = -2.29, R) >droEre2.scaffold_4770 2887333 101 + 17746568 CCCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU-GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAUGUCCUU ..........(((..........)))...((((((((..((.(((((.((((((((-(......)))))).......)))..))))).))..)))))))).. ( -30.71, z-score = -2.31, R) >droAna3.scaffold_13340 11326729 96 + 23697760 CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGGCGACAUGGGCCUGUCAUGUCUGCACUGGGCCUCUGACCCU----AUAUCC-- .((((((..((((.((..((((((((....)))).)))))).)))))))).))((.((((((((((....)).)).)))))).)).....----......-- ( -29.00, z-score = -0.86, R) >droWil1.scaffold_181130 9174324 88 + 16660200 -----GCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCAGCGACAUGGCCCUGUCAUG-------UACGCCUCUGGCCCACUAUGUGUCU-- -----.....(((.....((((((((....)))).))))((((((((..((((((((((...))))))-------).)))..)))))..)))....))).-- ( -28.90, z-score = -1.79, R) >consensus CUCCUGCAAUGACCCUAAUAACUGUCUACGGGCAUGUUAAGUGUCAGCGCCGACAU_GUCCUGUCAUGUCUGCACUGGGCCUCUGACCCUCCGCAUGUCCUU ......((..(.(((...((((((((....)))).))))(((((..(((..((((......)))).)))..)))))))).)..))................. (-20.25 = -21.13 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:34 2011