| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,740,186 – 6,740,271 |
| Length | 85 |
| Max. P | 0.774572 |

| Location | 6,740,186 – 6,740,271 |
|---|---|
| Length | 85 |
| Sequences | 12 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Shannon entropy | 0.34562 |
| G+C content | 0.44320 |
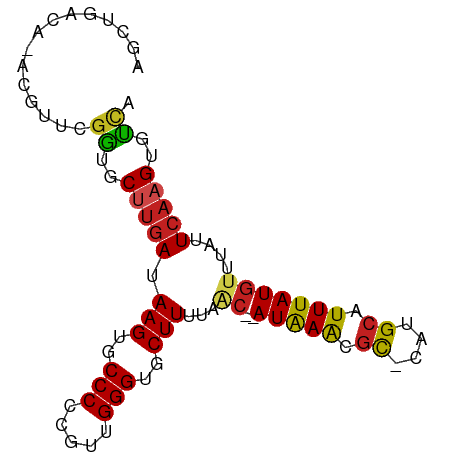
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774572 |
| Prediction | RNA |
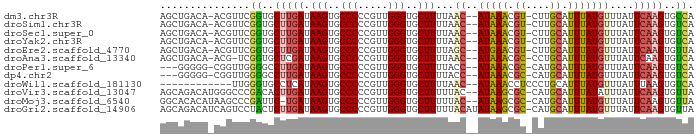
Download alignment: ClustalW | MAF
>dm3.chr3R 6740186 85 - 27905053 AGCUGACA-ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACGU-CUUGCAUUUAUGUUUAUUCAAGUGCCA ........-......((..(((((.(((..(((.....)))..)))..(((--(((((.(.-....).))))))))...)))))..)). ( -27.60, z-score = -3.15, R) >droSim1.chr3R 12899182 85 + 27517382 AGCUGACA-ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACGU-CUUGCAUUUAUGUUUAUUCAAGUGUCA ........-......((..(((((.(((..(((.....)))..)))..(((--(((((.(.-....).))))))))...)))))..)). ( -24.40, z-score = -2.26, R) >droSec1.super_0 5908759 85 + 21120651 AGCUGACA-ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACGU-CUUGCAUUUAUGUUUAUUCAAGUGUCA ........-......((..(((((.(((..(((.....)))..)))..(((--(((((.(.-....).))))))))...)))))..)). ( -24.40, z-score = -2.26, R) >droYak2.chr3R 10776232 85 - 28832112 AGCUGACA-ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACGU-CUUGCAUUUAUGUUUAUUCAAGUGUCA ........-......((..(((((.(((..(((.....)))..)))..(((--(((((.(.-....).))))))))...)))))..)). ( -24.40, z-score = -2.26, R) >droEre2.scaffold_4770 2878266 85 + 17746568 AGCUGACA-ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAGC--AUGAACGU-CUUGCAUUUAUGUUUAUUCAAGUGUUA ...(((((-(((((((.((((....(((..(((.....)))..)))..)))--))))))))-((((.((........)).))))))))) ( -26.40, z-score = -2.59, R) >droAna3.scaffold_13340 11318986 84 + 23697760 AGCUGACA-ACG-UCGGUGCUCGAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACGC-CCUGCAUUUAUGUUUAUUCAAGUGUCA ......((-(((-..((..((.....))..))..)))))((..(((..(((--(((((.((-...)).)))))))).....)))..)). ( -25.30, z-score = -2.35, R) >droPer1.super_6 3493272 82 - 6141320 ---GGGGG-CGGUUGGGGCCUUGAUAAGUGCCCCCGUUGGGUGCUUUUACC--AUAAACGC-CAUGCAUUUAUGUUUAUUCAAGUGUCA ---(((((-((.(((.........))).)))))))....((..(((....(--(((((.((-...)).)))))).......)))..)). ( -24.30, z-score = 0.28, R) >dp4.chr2 28159897 82 - 30794189 ---GGGGG-CGGUUGGGGCCUUGAUAAGUGCCCCCGUUGGGUGCUUUUACC--AUAAACGC-CAUGCAUUUAUGUUUAUUCAAGUGUCA ---(((((-((.(((.........))).)))))))....((..(((....(--(((((.((-...)).)))))).......)))..)). ( -24.30, z-score = 0.28, R) >droWil1.scaffold_181130 9198745 75 + 16660200 ------------UUGGGUGCCUCAUAAGUGCCCCCGUUGGGUGCUUUUAAC--AUAAACCUCCCUGCAUUUAUGUUUAUUUAAGUGUCA ------------..((((.......(((..(((.....)))..))).....--....))))....(((((((........))))))).. ( -17.59, z-score = -1.12, R) >droVir3.scaffold_13047 14056462 86 + 19223366 AGCAGACAUGGGCCCGACACUUGAUAAGUGCCCCCGUUGGGUGCUUUUUAC--AUAAGCGC-CAUGCAUUUAUAUUUAUUCAAGUGUUA .((........))..(((((((((((((((.....((..(((((((.....--..))))))-)..)).....)))))).))))))))). ( -24.30, z-score = -1.65, R) >droMoj3.scaffold_6540 21258370 85 - 34148556 GGCACACAUAAGCCCGAUUC-UGAUAAGUGCCCCCGUUGGGUGCUUUUUAC--AUAAGCGC-CAUGCAUUUAUGUUUAUUCAAGUGUUA (((((..((((((.......-..((((((((........(((((((.....--..))))))-)..))))))))))))))....))))). ( -19.80, z-score = -0.60, R) >droGri2.scaffold_14906 3603859 88 + 14172833 AGCAGACAUCAGUCCUACUCUUGAUAAGUGCCCCCGUUGGGUGCUUUUUACAUAUAAGCGC-CAUGCAUUUAUGUUUAUUCAAGUGUUA ....(((....)))..((.(((((.(((..(((.....)))..)))...(((((...((..-...))...)))))....))))).)).. ( -19.90, z-score = -1.27, R) >consensus AGCUGACA_ACGUUCGGUGCUUGAUAAGUGCCCCCGUUGGGUGCUUUUAAC__AUAAACGC_CAUGCAUUUAUGUUUAUUCAAGUGUCA ...............((..(((((.(((..(((.....)))..))).......(((((.((....)).)))))......)))))..)). (-16.76 = -16.32 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:30 2011