| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,731,777 – 6,731,879 |
| Length | 102 |
| Max. P | 0.655244 |

| Location | 6,731,777 – 6,731,879 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 60.93 |
| Shannon entropy | 0.75465 |
| G+C content | 0.43449 |
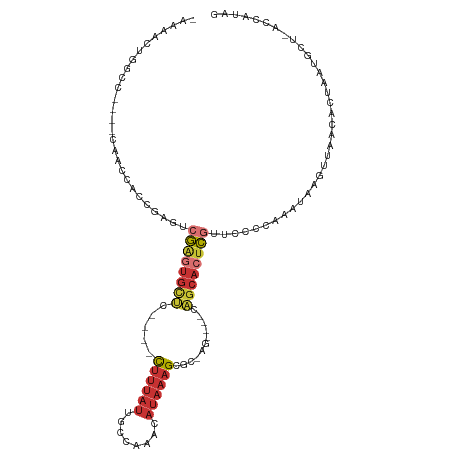
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -4.95 |
| Energy contribution | -5.32 |
| Covariance contribution | 0.37 |
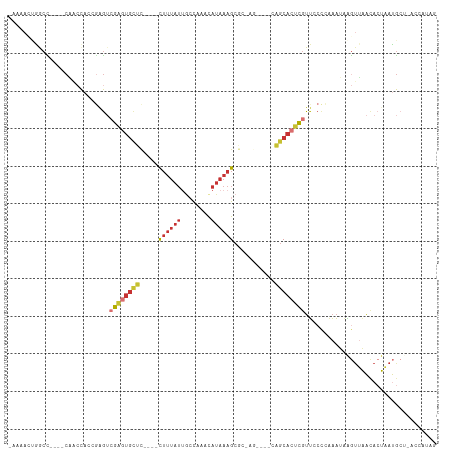
| Combinations/Pair | 1.50 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
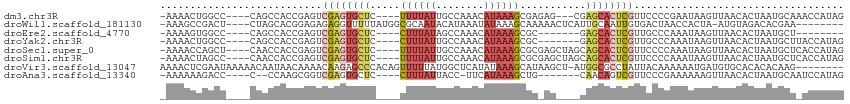
>dm3.chr3R 6731777 102 - 27905053 -AAAACUGGCC----CAGCCACCGAGUCGAGUGCUC----UUUUAUUGCCAAACAUAAAGCGAGAG---CGAGCACUCGUUCCCCGAAUAAGUUAACACUAAUGCAAACCAUAG -..(((((((.----..)))...(((.(((((((((----.(((((........)))))((....)---)))))))))))))........)))).................... ( -26.20, z-score = -3.39, R) >droWil1.scaffold_181130 9184120 100 + 16660200 -AAAGCCGACU----CUAGCACGGAGAGAGGUUUUUAUGGCGCAAUACAUAAAUAUAAAGCAAAAACUCAUUGCAAUUGUGACUAACCACUA-AUGUAGACACGAA-------- -.(((((..((----((.......)))).)))))(((((........))))).......((((.......))))..(((((.(((.......-...))).))))).-------- ( -16.70, z-score = -0.29, R) >droEre2.scaffold_4770 2869863 90 + 17746568 -AAAAGUGGCC----CAGCCACCGAGUCGAGUGCUC----CUUUAUAGCCAAACAUAAAGCGC-------GAGCACUCGUUGCCCAAAUAAGUUAACACUAAUGCU-------- -....(((((.----..))))).(...(((((((((----((((((........))))))...-------)))))))))...).......................-------- ( -24.10, z-score = -2.78, R) >droYak2.chr3R 10767654 98 - 28832112 -AAAACUGGCC----CAGCCACCGAGUCGAGUGCUC----CUUUAUUGCCAAACAUAAAGCGC-------GAGCACUCGUUGCCCAAAUAAGUUAACACUAAUGCUUACCAUAG -.....((((.----..))))..(...(((((((((----((((((........))))))...-------)))))))))...).....(((((..........)))))...... ( -22.20, z-score = -2.09, R) >droSec1.super_0 5900772 105 + 21120651 -AAAACCAGCU----CAACCACCGAGUCGAGUGCUC----UUUUAUUGCCAAACAUAAAGCGCGAGCUAGCAGCACUCGUUCCCCAAAUAAGUUAACACUAAUGCUCACCAUAG -..........----........(((((((((((((----(.((((........))))(((....))))).))))))))...........(((....)))...))))....... ( -20.20, z-score = -2.04, R) >droSim1.chr3R 12890904 105 + 27517382 -AAAACUAGCC----CAACCACCGAGUCGAGUGCUC----UUUUAUUGCCAAACAUAAAGCGCGAGCUAGCAGCACUCGUUCCCCAAAUAAGUUAACACUAAUGCUCACCAUAG -..........----........(((((((((((((----(.((((........))))(((....))))).))))))))...........(((....)))...))))....... ( -20.20, z-score = -2.16, R) >droVir3.scaffold_13047 5174669 105 - 19223366 AAAACUCGAAUAAAAACAAUAACAAAACAAGAGCCCACAGUUUUUAUGGCUCAUAUAAAGCAUAAGCU-AUGGCGCCUAUUACAAAAAAUGAUGUGCACACACAAG-------- .............................((.(((((.(((((.....(((.......)))..)))))-.))).))))..............((((....))))..-------- ( -14.50, z-score = -0.61, R) >droAna3.scaffold_13340 11311697 95 + 23697760 -AAAAAAGACC----C--CCAAGCGGUCGAGUGCUC----CUUUAUUACC-UUCAUAAAGCUG-------CAACAGUCGUUCCCGAAAAAAGUUAACACUAAUGCAAUCCAUAG -......((((----.--......))))...(((..----((((((....-...))))))..)-------))..(((.(((..(.......)..)))))).............. ( -12.90, z-score = -0.82, R) >consensus _AAAACUGGCC____CAACCACCGAGUCGAGUGCUC____CUUUAUUGCCAAACAUAAAGCGC_AG____CAGCACUCGUUCCCCAAAUAAGUUAACACUAAUGCU_ACCAUAG ...........................((((((((....................................))))))))................................... ( -4.95 = -5.32 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:29 2011