| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,728,374 – 6,728,477 |
| Length | 103 |
| Max. P | 0.915292 |

| Location | 6,728,374 – 6,728,477 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.33160 |
| G+C content | 0.32240 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.13 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
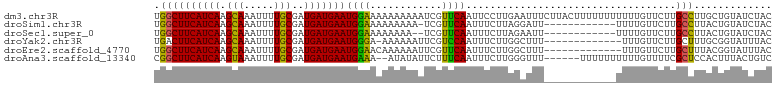
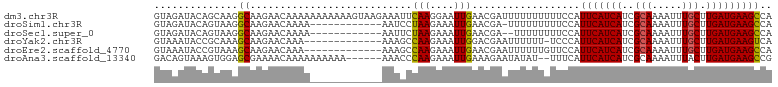
>dm3.chr3R 6728374 103 + 27905053 UGGCUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGAAAAAAAAAAUCGUUCAAUUCCUUGAAUUUCUUACUUUUUUUUUUUGUUCUUGCCUUGCUGUAUCUAC .(((.(((((..(((.....)))))))).(((..((((((((((...((((((....)))))).......))))))))))..)))..)))............. ( -22.20, z-score = -2.57, R) >droSim1.chr3R 12887503 90 - 27517382 UGGCUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGAAAAAAAAA-UCGUUCAAUUUCUUAGGAUU------------UUUUGUUCUUGCCUUACUGUAUCUAC .((((((((((.(((.....)))..))))))).((((.((((((-((.(..........).))))------------)))).)))).)))............. ( -20.90, z-score = -2.52, R) >droSec1.super_0 5897383 89 - 21120651 UGGCUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGAAAAAAAA--UCGUUCAAUUUCUUAGAAUU------------UUUUGUUCUUGCCUUACUGUAUCUAC .((((((((((.(((.....)))..))))))).((((...(((--..((((.........)))).------------.))).)))).)))............. ( -17.30, z-score = -1.47, R) >droYak2.chr3R 10764198 89 + 28832112 UGACUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGGA-AAAAAAUUCGUCCAAUUUCUUGGCUUU-------------UUUGUUCUUGCUUUGCGGUAUUUAC ....(((((((.(((.....)))..))))))).((((-(.((((...(.((((....)))))..)-------------))).)))))................ ( -15.20, z-score = 0.21, R) >droEre2.scaffold_4770 2866583 90 - 17746568 UGGCUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGAACAAAAAAUUCGUUCAAUUUCUUGGCUUU-------------UUUGUUCUUGCUUUACGGUAUUUAC .((((((((((.(((.....)))..))))))).(((((((((((..((..........))..)))-------------)))))))).)))............. ( -21.40, z-score = -2.55, R) >droAna3.scaffold_13340 11308390 95 - 23697760 CGGCUUCAUCAAGUAAAUUUUGCGAUGAUGAAUGAAA--AUAUAUUCUUUCAAUUUCUUGGGUUU------UUUUUUUUUUGUUUUCGCUCCACUUUACUGUC ...........((((((....((((.((..((.((((--(...((((............))))..------.))))).))..)).)))).....))))))... ( -15.20, z-score = -1.04, R) >consensus UGGCUUCAUCAAGCAAAUUUUGCGAUGAUGAAUGGAAAAAAAAAUUCGUUCAAUUUCUUGGAUUU____________UUUUGUUCUUGCCUUACUGUAUCUAC .((((((((((.(((.....)))..)))))))((((............))))...................................)))............. (-11.77 = -11.13 + -0.64)
| Location | 6,728,374 – 6,728,477 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.33160 |
| G+C content | 0.32240 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -10.71 |
| Energy contribution | -10.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
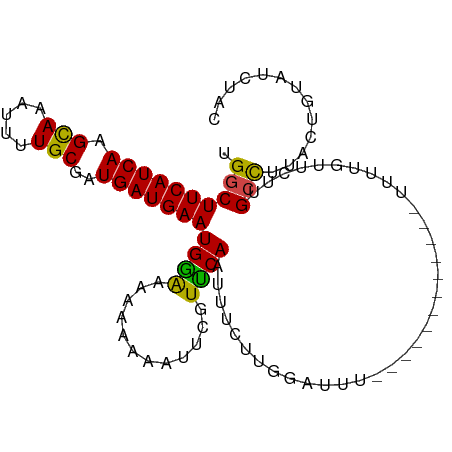
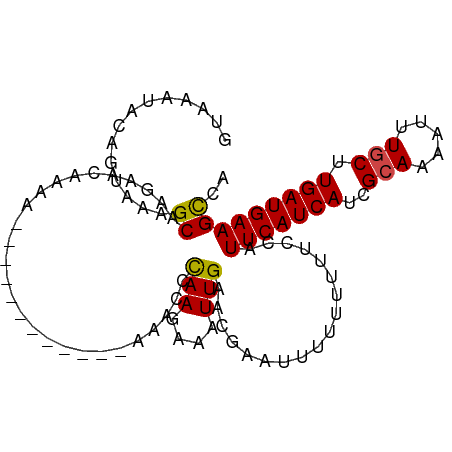
>dm3.chr3R 6728374 103 - 27905053 GUAGAUACAGCAAGGCAAGAACAAAAAAAAAAAGUAAGAAAUUCAAGGAAUUGAACGAUUUUUUUUUUCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGCCA .............(((........(((((((((((....(((((...))))).....)))))))))))...(((((((..(((.....))).)))))))))). ( -21.80, z-score = -2.27, R) >droSim1.chr3R 12887503 90 + 27517382 GUAGAUACAGUAAGGCAAGAACAAAA------------AAUCCUAAGAAAUUGAACGA-UUUUUUUUUCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGCCA .............(((..(((.((((------------((((.(((....)))...))-)))))).)))..(((((((..(((.....))).)))))))))). ( -21.80, z-score = -3.61, R) >droSec1.super_0 5897383 89 + 21120651 GUAGAUACAGUAAGGCAAGAACAAAA------------AAUUCUAAGAAAUUGAACGA--UUUUUUUUCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGCCA .............(((..(((.((((------------((((.(((....)))...))--)))))))))..(((((((..(((.....))).)))))))))). ( -18.80, z-score = -2.38, R) >droYak2.chr3R 10764198 89 - 28832112 GUAAAUACCGCAAAGCAAGAACAAA-------------AAAGCCAAGAAAUUGGACGAAUUUUUU-UCCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGUCA ..................(((.(((-------------(..(((((....)))).)...)))).)-))...(((((((..(((.....))).))))))).... ( -16.10, z-score = -1.42, R) >droEre2.scaffold_4770 2866583 90 + 17746568 GUAAAUACCGUAAAGCAAGAACAAA-------------AAAGCCAAGAAAUUGAACGAAUUUUUUGUUCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGCCA ..............((..(((((((-------------(((..(((....)))......))))))))))..(((((((..(((.....))).))))))))).. ( -20.70, z-score = -3.57, R) >droAna3.scaffold_13340 11308390 95 + 23697760 GACAGUAAAGUGGAGCGAAAACAAAAAAAAAA------AAACCCAAGAAAUUGAAAGAAUAUAU--UUUCAUUCAUCAUCGCAAAAUUUACUUGAUGAAGCCG ..((...(((((((((((..............------.....(((....)))...((((....--....))))....))))....)))))))..))...... ( -13.20, z-score = -1.51, R) >consensus GUAAAUACAGUAAAGCAAGAACAAAA____________AAACCCAAGAAAUUGAACGAAUUUUUUUUUCCAUUCAUCAUCGCAAAAUUUGCUUGAUGAAGCCA ..............((...........................(((....)))..................(((((((..(((.....))).))))))))).. (-10.71 = -10.52 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:29 2011