| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,678,100 – 6,678,205 |
| Length | 105 |
| Max. P | 0.994385 |

| Location | 6,678,100 – 6,678,205 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.68 |
| Shannon entropy | 0.59510 |
| G+C content | 0.53357 |
| Mean single sequence MFE | -38.06 |
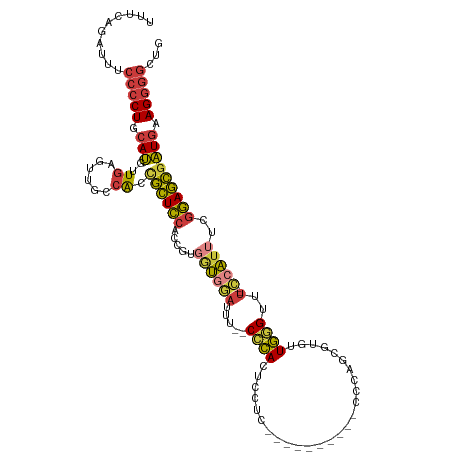
| Consensus MFE | -21.05 |
| Energy contribution | -21.04 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
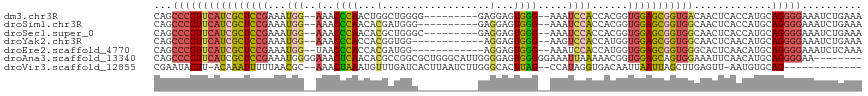
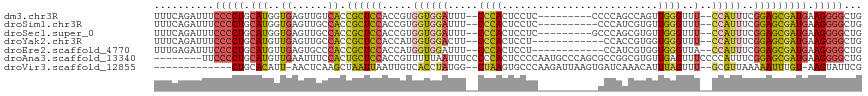
>dm3.chr3R 6678100 105 + 27905053 CAGCCCCUUCAUCGCUCCGAAAUGG--AAACCCAACUGGCUGGGG---------GAGGAGUGGG--AAAUCCACCACGGUGGAGCGGUGACAACUCACCAUGCAGGGGAAAUCUGAAA ((((((((.(((.((((((...(((--...((((......)))).---------..(((.....--...))).)))...))))))(((((....)))))))).)))))....)))... ( -41.10, z-score = -1.70, R) >droSim1.chr3R 12829042 104 - 27517382 CAGCCCCUUCAUCGCUCCGAAAUGG--AAACCCAACACGAUGGG----------GAGGAGUGGG--AAAUCCACCACGGUGGAGCGGUGGCAACUCACCAUGCAGGGGAAAUCUGAAA ((((((((.((((((((((...(((--...((((......))))----------..(((.....--...))).)))...))))))))))(((........))))))))....)))... ( -40.90, z-score = -2.19, R) >droSec1.super_0 5847825 105 - 21120651 CAGCCCCUUCAUCGCUCCGAAAUGG--AAACCCAACACGCUGGGC---------GAGGAGUGGG--AAAUCCACCACGGUGGAGCGGUGGCAACUCACCAUGCAGGGGAAAUCUGAAA ((((((((.((((((((((...(((--...((((......)))).---------..(((.....--...))).)))...))))))))))(((........))))))))....)))... ( -40.20, z-score = -1.69, R) >droYak2.chr3R 10712423 102 + 28832112 CAGCCCCUUCAUCGCUCCGAAAUGG--AAACCCACCACGGUGG------------AGGAGUGGG--AAGUCCACCAUGGUGGAGCGGUGGCAACUCAACAUGCAGGGGAAAUCUGAAA ((((((((.((((((((((..((((--....((((....))))------------.(((.(...--.).))).))))..))))))))))(((........))))))))....)))... ( -45.40, z-score = -3.26, R) >droEre2.scaffold_4770 2813741 102 - 17746568 CAGCCCCUUCAUCGCUCCGAAAUGG--UAACCCACCACGAUGG------------AGGAGUGGG--AAAUCCACCAUGGUGGAGCGGUGGGCACUCAACAUGCAGGGGAAAUCUCAAA ...((((((((((((((((..((((--(..(((((..(.....------------.)..)))))--......)))))..)))))))))))(((.......)))))))).......... ( -43.60, z-score = -3.03, R) >droAna3.scaffold_13340 6995729 110 + 23697760 CAGCCCCUUCAUCGCUCCGAAAUGGGGAAACUCAACACGCCGGCGCUGGGCAUUGGGGAGUGGGGGAAAUUAAAAACGGUGGAGCAGUGGAAAUUCAACAUGCAGGGGAA-------- ...(((((((.(((((((....((((....))))....(((.......))).....)))))))..)))...............(((.(((....)))...))).))))..-------- ( -35.10, z-score = -0.60, R) >droVir3.scaffold_12855 130165 99 + 10161210 CGAAUACUU-ACAAAUUUUUAACGC--AAACUAAAUGUUUGAUCACUUAAUCUUGGGCACUUAG--CCAUAGGUGACAAUUAAUUAGCUUGAGUU-AAUGUGCAG------------- ........(-(((.....(((((.(--((.((((.((.(((.(((((((......(((.....)--)).)))))))))).)).)))).))).)))-))))))...------------- ( -20.10, z-score = -1.52, R) >consensus CAGCCCCUUCAUCGCUCCGAAAUGG__AAACCCAACACGCUGGG__________GAGGAGUGGG__AAAUCCACCACGGUGGAGCGGUGGCAACUCAACAUGCAGGGGAAAUCUGAAA ...((((((((((((((((...(((.....((((..........................))))......)))......))))))))))).............))))).......... (-21.05 = -21.04 + -0.01)
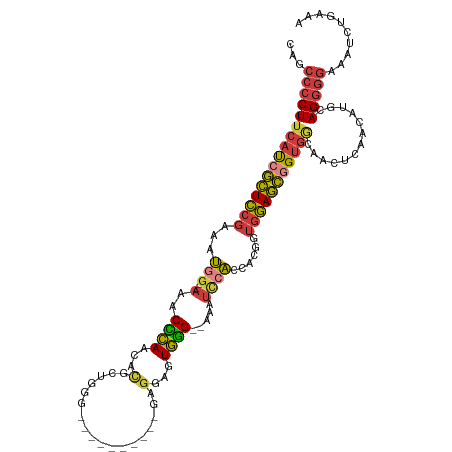
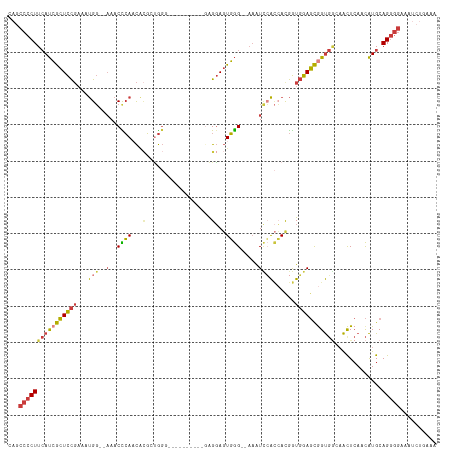
| Location | 6,678,100 – 6,678,205 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.68 |
| Shannon entropy | 0.59510 |
| G+C content | 0.53357 |
| Mean single sequence MFE | -35.71 |
| Consensus MFE | -18.89 |
| Energy contribution | -20.19 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6678100 105 - 27905053 UUUCAGAUUUCCCCUGCAUGGUGAGUUGUCACCGCUCCACCGUGGUGGAUUU--CCCACUCCUC---------CCCCAGCCAGUUGGGUUU--CCAUUUCGGAGCGAUGAAGGGGCUG ...(((....(((((.((((((((....)))))(((((...(..((((....--.))))..)..---------.(((((....)))))...--.......))))).))).)))))))) ( -40.40, z-score = -2.25, R) >droSim1.chr3R 12829042 104 + 27517382 UUUCAGAUUUCCCCUGCAUGGUGAGUUGCCACCGCUCCACCGUGGUGGAUUU--CCCACUCCUC----------CCCAUCGUGUUGGGUUU--CCAUUUCGGAGCGAUGAAGGGGCUG ...(((....(((((.(((((((......))))(((((...(..((((....--.))))..)..----------((((......))))...--.......))))).))).)))))))) ( -38.70, z-score = -1.92, R) >droSec1.super_0 5847825 105 + 21120651 UUUCAGAUUUCCCCUGCAUGGUGAGUUGCCACCGCUCCACCGUGGUGGAUUU--CCCACUCCUC---------GCCCAGCGUGUUGGGUUU--CCAUUUCGGAGCGAUGAAGGGGCUG ...(((....(((((.(((((((......))))(((((...(..((((....--.))))..)..---------((((((....))))))..--.......))))).))).)))))))) ( -41.00, z-score = -2.03, R) >droYak2.chr3R 10712423 102 - 28832112 UUUCAGAUUUCCCCUGCAUGUUGAGUUGCCACCGCUCCACCAUGGUGGACUU--CCCACUCCU------------CCACCGUGGUGGGUUU--CCAUUUCGGAGCGAUGAAGGGGCUG ...(((....((((((((........)))((.(((((((((((((((((...--........)------------))))))))))((....--)).....)))))).)).)))))))) ( -42.20, z-score = -3.04, R) >droEre2.scaffold_4770 2813741 102 + 17746568 UUUGAGAUUUCCCCUGCAUGUUGAGUGCCCACCGCUCCACCAUGGUGGAUUU--CCCACUCCU------------CCAUCGUGGUGGGUUA--CCAUUUCGGAGCGAUGAAGGGGCUG ..........(((((((((.....)))).((.(((((((((((((((((...--........)------------))))))))))((....--)).....)))))).)).)))))... ( -41.80, z-score = -2.91, R) >droAna3.scaffold_13340 6995729 110 - 23697760 --------UUCCCCUGCAUGUUGAAUUUCCACUGCUCCACCGUUUUUAAUUUCCCCCACUCCCCAAUGCCCAGCGCCGGCGUGUUGAGUUUCCCCAUUUCGGAGCGAUGAAGGGGCUG --------..((((((((.((.(.....).)))))..((.(((((..((........((((....(((((.......)))))...))))........))..))))).)).)))))... ( -25.59, z-score = -0.19, R) >droVir3.scaffold_12855 130165 99 - 10161210 -------------CUGCACAUU-AACUCAAGCUAAUUAAUUGUCACCUAUGG--CUAAGUGCCCAAGAUUAAGUGAUCAAACAUUUAGUUU--GCGUUAAAAAUUUGU-AAGUAUUCG -------------.((((..((-(((.((((((((....(((((((.((.((--(.....)))......)).)))).)))....)))))))--).))))).....)))-)........ ( -20.30, z-score = -1.96, R) >consensus UUUCAGAUUUCCCCUGCAUGUUGAGUUGCCACCGCUCCACCGUGGUGGAUUU__CCCACUCCUC__________CCCAGCGUGUUGGGUUU__CCAUUUCGGAGCGAUGAAGGGGCUG ..........(((((.(((..((......)).((((((.....(((((.......)))))...............((((....)))).............))))))))).)))))... (-18.89 = -20.19 + 1.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:25 2011