
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,676,595 – 6,676,661 |
| Length | 66 |
| Max. P | 0.810645 |

| Location | 6,676,595 – 6,676,661 |
|---|---|
| Length | 66 |
| Sequences | 9 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 61.56 |
| Shannon entropy | 0.76342 |
| G+C content | 0.48704 |
| Mean single sequence MFE | -14.77 |
| Consensus MFE | -5.85 |
| Energy contribution | -5.70 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6676595 66 + 27905053 CUGGCACCAGAAAGUCCUGGCUCCAUCGAGGCGCGACGGAAUUAUUC--CUUAAAUACAUAUACAUAU---- (((....)))...(((.((.(((....))).)).)))(((.....))--)..................---- ( -13.20, z-score = -0.65, R) >droEre2.scaffold_4770 2812184 60 - 17746568 CUGGCACCAGAAAGUCCUGGCUCCAUCGAGGCGCGACGGAAUUAUUC--CUUAAACACAUAU---------- (((....)))...(((.((.(((....))).)).)))(((.....))--)............---------- ( -13.20, z-score = -0.70, R) >droYak2.chr3R 10710792 60 + 28832112 CUGGCACCAGAAAGUCCUGGCUCCAUAGAGGCGCGACGGAAUUAUUC--CUUAAAUACAUAU---------- (((....)))...(((.((.(((....))).)).)))(((.....))--)............---------- ( -13.30, z-score = -0.85, R) >droSec1.super_0 5846371 60 - 21120651 CUGGCACCAGAAAGUCCUGGCUCCAUCGAGGCGCGACGGAAUUAUUC--CUUAAAUACAUAU---------- (((....)))...(((.((.(((....))).)).)))(((.....))--)............---------- ( -13.20, z-score = -0.67, R) >droSim1.chr3R 12827607 60 - 27517382 CUGGCACCAGAAAGUCCUGGCUCCAUCGAGGCGCGACGGAAUUAUUC--CUUAAAUACAUAU---------- (((....)))...(((.((.(((....))).)).)))(((.....))--)............---------- ( -13.20, z-score = -0.67, R) >droPer1.super_3 7312345 51 - 7375914 ---------------CUUGGCUCCGCCUAGACGGAAUGAAAUCAUUG--UUUAGUUUUCUAGGCAUAU---- ---------------.........(((((((..((((.((((....)--))).)))))))))))....---- ( -12.30, z-score = -1.50, R) >droVir3.scaffold_12855 127928 66 + 10161210 CCGGCUCCAGCAAGUCCCGGCUCCACUCGGGCGCGACAGUAUUACGA----UGUUUUACUACUUGUGCUA-- ((((..(......)..)))).........(((((((.((((..((..----.))..))))..))))))).-- ( -15.10, z-score = 0.62, R) >droMoj3.scaffold_6540 2209653 65 + 34148556 --GGCUCCAGCUCGUCUCGGCUCCACUCAGGCGAGACAGCAUAAUGA----UGUUUUACUAUUUGUGCUGC- --(((....))).((((((.((......)).))))))(((((((...----.(.....)...)))))))..- ( -19.30, z-score = -1.20, R) >droGri2.scaffold_14906 13186747 72 + 14172833 CUGGGUCCAGCUUGUUCCGGCUCCACUGGGGCGAGACAAUUUGACUAAGUAUGUUUUACUACUUGUCUCUCC (((....)))((((((((((.....)))))))))).......(((.(((((........))))))))..... ( -20.10, z-score = -1.08, R) >consensus CUGGCACCAGAAAGUCCUGGCUCCAUCGAGGCGCGACGGAAUUAUUC__CUUAAAUACAUAU___U______ .............(((.((.((......)).)).)))................................... ( -5.85 = -5.70 + -0.15)

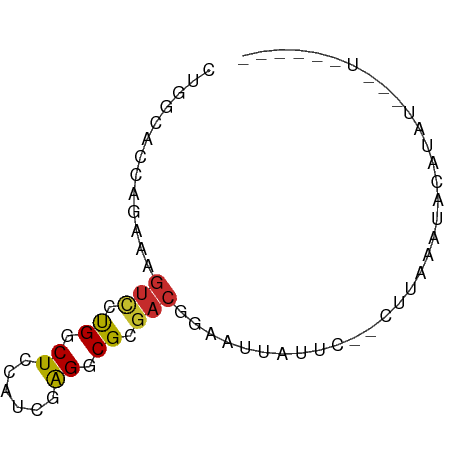

| Location | 6,676,595 – 6,676,661 |
|---|---|
| Length | 66 |
| Sequences | 9 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 61.56 |
| Shannon entropy | 0.76342 |
| G+C content | 0.48704 |
| Mean single sequence MFE | -15.97 |
| Consensus MFE | -6.50 |
| Energy contribution | -7.24 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6676595 66 - 27905053 ----AUAUGUAUAUGUAUUUAAG--GAAUAAUUCCGUCGCGCCUCGAUGGAGCCAGGACUUUCUGGUGCCAG ----..................(--(......(((((((.....)))))))((((((....)))))).)).. ( -17.30, z-score = -1.09, R) >droEre2.scaffold_4770 2812184 60 + 17746568 ----------AUAUGUGUUUAAG--GAAUAAUUCCGUCGCGCCUCGAUGGAGCCAGGACUUUCUGGUGCCAG ----------............(--(......(((((((.....)))))))((((((....)))))).)).. ( -17.30, z-score = -1.28, R) >droYak2.chr3R 10710792 60 - 28832112 ----------AUAUGUAUUUAAG--GAAUAAUUCCGUCGCGCCUCUAUGGAGCCAGGACUUUCUGGUGCCAG ----------............(--((.....)))............(((.((((((....)))))).))). ( -13.40, z-score = -0.28, R) >droSec1.super_0 5846371 60 + 21120651 ----------AUAUGUAUUUAAG--GAAUAAUUCCGUCGCGCCUCGAUGGAGCCAGGACUUUCUGGUGCCAG ----------............(--(......(((((((.....)))))))((((((....)))))).)).. ( -17.30, z-score = -1.51, R) >droSim1.chr3R 12827607 60 + 27517382 ----------AUAUGUAUUUAAG--GAAUAAUUCCGUCGCGCCUCGAUGGAGCCAGGACUUUCUGGUGCCAG ----------............(--(......(((((((.....)))))))((((((....)))))).)).. ( -17.30, z-score = -1.51, R) >droPer1.super_3 7312345 51 + 7375914 ----AUAUGCCUAGAAAACUAAA--CAAUGAUUUCAUUCCGUCUAGGCGGAGCCAAG--------------- ----...((((((((........--.((((....))))...))))))))........--------------- ( -11.20, z-score = -2.05, R) >droVir3.scaffold_12855 127928 66 - 10161210 --UAGCACAAGUAGUAAAACA----UCGUAAUACUGUCGCGCCCGAGUGGAGCCGGGACUUGCUGGAGCCGG --..(((((.(((.((.....----...)).)))))).)).......(((..((((......))))..))). ( -13.30, z-score = 1.67, R) >droMoj3.scaffold_6540 2209653 65 - 34148556 -GCAGCACAAAUAGUAAAACA----UCAUUAUGCUGUCUCGCCUGAGUGGAGCCGAGACGAGCUGGAGCC-- -.((((....(((((......----..)))))..(((((((.((......)).))))))).)))).....-- ( -16.30, z-score = -0.37, R) >droGri2.scaffold_14906 13186747 72 - 14172833 GGAGAGACAAGUAGUAAAACAUACUUAGUCAAAUUGUCUCGCCCCAGUGGAGCCGGAACAAGCUGGACCCAG ((.(((((((..((((.....))))........))))))).))....(((..((((......))))..))). ( -20.30, z-score = -2.19, R) >consensus ______A___AUAUGUAUUUAAG__GAAUAAUUCCGUCGCGCCUCGAUGGAGCCAGGACUUUCUGGUGCCAG ...............................................(((..((((......))))..))). ( -6.50 = -7.24 + 0.74)
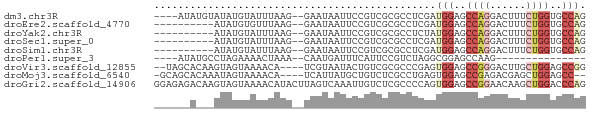

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:23 2011