
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,666,829 – 6,666,941 |
| Length | 112 |
| Max. P | 0.911620 |

| Location | 6,666,829 – 6,666,941 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.90 |
| Shannon entropy | 0.48348 |
| G+C content | 0.45073 |
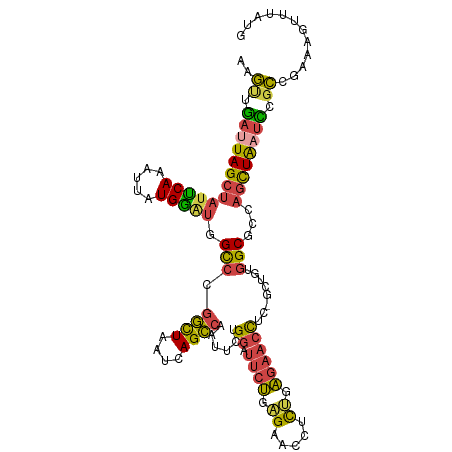
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -19.58 |
| Energy contribution | -18.95 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
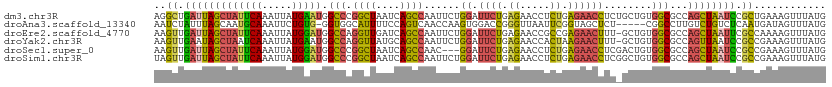
>dm3.chr3R 6666829 112 + 27905053 AGGCUGAUUAGCUAUUCAAAUUAUGAAUGGCCCGGCUAAUCAGCCAAUUCUGGAUUCUGAGAACCUCUGAGAACCUCUGCUGUGGCGCCAGCUAAUCCGCUGAAAGUUUAUG .(((.(((((((((((((.....)))))(((((((((....))))......((.((((.((.....)).))))))........)).))))))))))).)))........... ( -37.50, z-score = -3.10, R) >droAna3.scaffold_13340 6984648 106 + 23697760 AAUCUAUUUAGCAAUGCAAAUUCUGUG-GGUGGCAUUUUCCAGUCAACCAAGUGGACCGGGUUAAUUCGGUAGCUCU-----CGGCCUUGUCUGUCUCAAUGAUAGUUUAUG ..........((((.((........((-(.((((........)))).)))(((..((((((....)))))).)))..-----..)).))))(((((.....)))))...... ( -22.80, z-score = 0.20, R) >droEre2.scaffold_4770 2802323 111 - 17746568 AAGUUGAUUAGCUAUUCAAAUUAUGGAUGGCCAGGUUGAUCAGCCAAUUCUGGAUUCUGAGAACCGCCGAGAACUUU-GCUGUGGCGCCAGCUAAUUCGCCAAAAGUUUAUG ..((.(((((((((((((.....)))))(((..((((..(((((((....)))...)))).))))((((((......-.)).))))))))))))))).))............ ( -30.70, z-score = -1.10, R) >droYak2.chr3R 10700859 111 + 28832112 AAGUUGAAUAGCUAAUCAAAUUAUGAAUGGCCAGGUUAUGCAGCCAAUUCUGGAUUCUGAGAACCACUAAGAACUUU-GCUGUGGCGCCAGUUAAUCCGCCGAAAGUUUAUG .(((......((((.(((.....))).))))..((((...((((((....)))...)))..)))))))..(((((((-....(((((..........))))))))))))... ( -22.80, z-score = 0.36, R) >droSec1.super_0 5836540 109 - 21120651 AAGUUGAUUAGCUAUUCAAAUUAUGGAUGGCCCGGCUAAUCAGCCAAC---GGAUUCUGAGAACCUCUGAGAACCUCGACUGUGGCGCCAGCUAAUCCGCCGAAAGUUUAUG ..((.(((((((((((((.....)))))(((((((((....))))...---((.((((.((.....)).))))))........)).))))))))))).))(....)...... ( -34.60, z-score = -2.87, R) >droSim1.chr3R 12817405 112 - 27517382 UAGUUGAUUAGCUAUUCAAAUUAUGGAUGGCCCGGCUAAUCAGCCAAUUCUGGAUUCUGAGAACCUCUGAGAACCUCGGCUGUGGCGCCAGCUAAUCCGCCGAAAGUUUAUG ..((.(((((((((((((.....)))))(((...((((..(((((......((.((((.((.....)).))))))..)))))))))))))))))))).))(....)...... ( -35.00, z-score = -2.06, R) >consensus AAGUUGAUUAGCUAUUCAAAUUAUGGAUGGCCCGGCUAAUCAGCCAAUUCUGGAUUCUGAGAACCUCUGAGAACCUC_GCUGUGGCGCCAGCUAAUCCGCCGAAAGUUUAUG ..((.(((((((((((((.....))))).(((.((((....))))......((.((((.((.....)).))))))........)))...)))))))).))............ (-19.58 = -18.95 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:21 2011