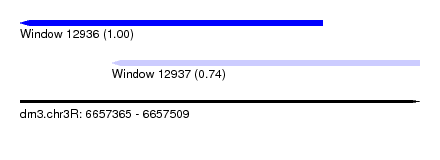
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,657,365 – 6,657,509 |
| Length | 144 |
| Max. P | 0.996925 |

| Location | 6,657,365 – 6,657,474 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.79 |
| Shannon entropy | 0.50243 |
| G+C content | 0.39309 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6657365 109 - 27905053 UGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGCGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAAGUAAUGUGUAUGCUUUAAAGUGUCUCGAAGGCUUGCCUUUCA (((((.((.((((((((((.((..--.....)).)))))))))).)).)))))..(((((((((((((..(((.((....)).)))...))))))(....)..))))))). ( -34.80, z-score = -2.90, R) >droSim1.chr3R 12807988 108 + 27517382 UGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUUAAAGUGUCUCGAAGGCAAGCCUUUCA (((((.((.((((((((((.((..--.....)).)))))))))).)).)))))..(((((((((((((.-(((.((....)).)))...))))))(....)..))))))). ( -34.00, z-score = -2.82, R) >droYak2.chr3R 10691068 108 - 28832112 UGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUCAAAGUGUCUCGAAGUCUCGCCUUUUA (((((.((.((((((((((.((..--.....)).)))))))))).)).)))))..((((((((((((..-...((.(.(..((....))..)).))..)))))))))))). ( -34.40, z-score = -3.34, R) >droEre2.scaffold_4770 2792968 108 + 17746568 UGGUGCUUUCUAAGAAUUUUCUUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGUGUGCUUUAAAGUGUCUCGAAGUCUCGCCUUUUA (((((.((.((((((((((.(...--......).)))))))))).)).)))))..((((((((((((..-...((.(.(..((....))..)).))..)))))))))))). ( -32.70, z-score = -3.11, R) >droWil1.scaffold_181130 5893727 106 - 16660200 CGUUUCGUCUUUAUAUUUAUUAUCUGGCGCCAAUUAAACUACAGCGGUCUACAAAAGACUUCAAAAUA--UGGCUCAU---CUUUAAAAGAAAUGUGUCAUUCACAAAGCA .((((((((...(((.....)))..))))......))))....(((((((.....))))).......(--((((.(((---.((.....)).))).))))).......)). ( -13.90, z-score = 0.02, R) >consensus UGGUGCUUUCUAAGAAUUUUCAUU__GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA_UAAUGUGUGUGCUUUAAAGUGUCUCGAAGGCUCGCCUUUCA .((((.(((((((((((((.((.........)).)))))))))).)))))))...(((((((((((((..(((.((....)).)))...))))))........))))))). (-18.92 = -19.64 + 0.72)


| Location | 6,657,398 – 6,657,509 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.46483 |
| G+C content | 0.37085 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -14.24 |
| Energy contribution | -13.24 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6657398 111 - 27905053 --UUCCUAUCAG-CAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGCGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAAGUAAUGUGU --.((((.((((-((......))))))........((((((((.((.((((((((((.((..--.....)).)))))))))).)).)))))).))))))................. ( -27.70, z-score = -1.71, R) >droSim1.chr3R 12808021 110 + 27517382 --UUCCUAUCAG-CAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGU --.((((.((((-((......))))))........((((((((.((.((((((((((.((..--.....)).)))))))))).)).)))))).))))))........-........ ( -27.70, z-score = -1.99, R) >droYak2.chr3R 10691101 110 - 28832112 --UUCCUAUCUG-CAAAUCAAUGUCGGCUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGU --.........(-((......((((.((((.....((((((((.((.((((((((((.((..--.....)).)))))))))).)).)))))).)).))))..)))).-.....))) ( -28.30, z-score = -2.00, R) >droEre2.scaffold_4770 2793001 110 + 17746568 --UUCCUAUCUG-CAAAUCAAUGUUGACUCAAAUACCGUGGUGCUUUCUAAGAAUUUUCUUU--GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA-UAAUGUGU --..........-.....((.(((((.(((.....((((((((.((.((((((((((.(...--......).)))))))))).)).)))).)))).....))).)))-)).))... ( -25.70, z-score = -1.35, R) >droWil1.scaffold_181130 5893759 112 - 16660200 UUUAUACAUUAAUCACAUCUCAUUUGGGUUAAAUGUUUCGUUUCGUCUUUAUAUUUAUUAUCUGGCGCCAAUUAAACUACAGCGGUCUACAAAAGACUUCAAAAUA--UGGCUC-- .........................((((((...((((.(((.((((...(((.....)))..))))..))).))))....(.(((((.....))))).)......--))))))-- ( -12.90, z-score = 0.10, R) >consensus __UUCCUAUCAG_CAAAUCAAUGUUGACUCAAAUACCUUGGUGCUUUCUAAGAAUUUUCAUU__GUACUUGUGAAUUCUUGGCAAAUACCAACGGAGGGCGAGACAA_UAAUGUGU ...........................(((.....((((((((.(((((((((((((.((.........)).)))))))))).))))))))).)).....)))............. (-14.24 = -13.24 + -1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:20 2011