
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,583,002 – 6,583,145 |
| Length | 143 |
| Max. P | 0.983047 |

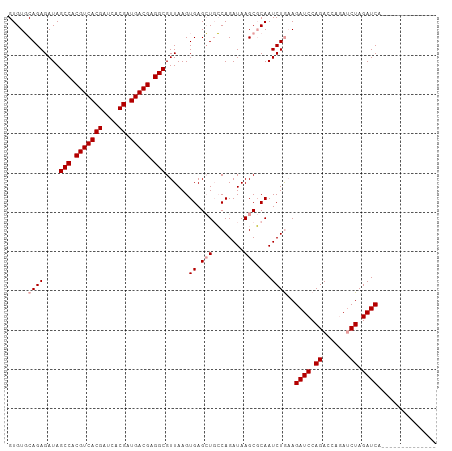
| Location | 6,583,002 – 6,583,111 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Shannon entropy | 0.20219 |
| G+C content | 0.49281 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6583002 109 + 27905053 GUGUUCAGAGAUAGCCACGUCACGAUCACGAUGACGAGGCGUUAAGUGAGAUUGCAGAUAAGCCCAAUCUACAGAUCCAGACCAUAACUAGAUCUGCUAUCUCCAAGCA .((((..(((((((((.(((((((....)).))))).)))........((((((..........)))))).((((((.((.......)).)))))).))))))..)))) ( -32.30, z-score = -3.18, R) >droSim1.chr3R 12739222 95 - 27517382 GUGUGCAGAGAUAGCCACGUCACGAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCGCAAUCUGAAGAUCCAGACCAGAUCUAGAUCA-------------- .....((((....(((.(((((((....)).))))).)))......((.(((........))).)).))))..((((.(((.....))).)))).-------------- ( -27.20, z-score = -1.52, R) >droSec1.super_0 5761749 95 - 21120651 GUGUGCAGAGAUAGCCACGUCACGAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCGCAAUCUGAAGAUCCAGACCAGAUCUAGAUCA-------------- .....((((....(((.(((((((....)).))))).)))......((.(((........))).)).))))..((((.(((.....))).)))).-------------- ( -27.20, z-score = -1.52, R) >consensus GUGUGCAGAGAUAGCCACGUCACGAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCGCAAUCUGAAGAUCCAGACCAGAUCUAGAUCA______________ .....((((....(((.(((((((....)).))))).)))......((.(((........))).)).))))..((((.((.......)).))))............... (-21.70 = -22.37 + 0.67)


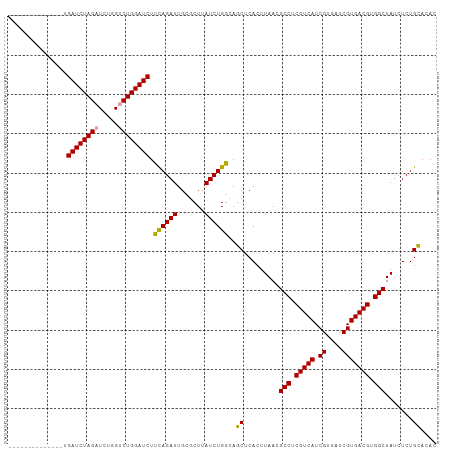
| Location | 6,583,002 – 6,583,111 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.66 |
| Shannon entropy | 0.20219 |
| G+C content | 0.49281 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -28.33 |
| Energy contribution | -27.56 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6583002 109 - 27905053 UGCUUGGAGAUAGCAGAUCUAGUUAUGGUCUGGAUCUGUAGAUUGGGCUUAUCUGCAAUCUCACUUAACGCCUCGUCAUCGUGAUCGUGACGUGGCUAUCUCUGAACAC ((.(..((((((((((((((((.......))))))))))((((((.(......).))))))........(((.(((((.((....))))))).)))))))))..).)). ( -41.60, z-score = -3.92, R) >droSim1.chr3R 12739222 95 + 27517382 --------------UGAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGGCAGCUCACUUAACGCCUCGUCAUCGUGAUCGUGACGUGGCUAUCUCUGCACAC --------------.(((((((((...)))))))))..((((..(((((.....))).)).........(((.(((((.((....))))))).)))....))))..... ( -30.80, z-score = -2.06, R) >droSec1.super_0 5761749 95 + 21120651 --------------UGAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGGCAGCUCACUUAACGCCUCGUCAUCGUGAUCGUGACGUGGCUAUCUCUGCACAC --------------.(((((((((...)))))))))..((((..(((((.....))).)).........(((.(((((.((....))))))).)))....))))..... ( -30.80, z-score = -2.06, R) >consensus ______________UGAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGGCAGCUCACUUAACGCCUCGUCAUCGUGAUCGUGACGUGGCUAUCUCUGCACAC ...............((((((((.....)))))))).((((((.......))))))..((.........(((.(((((.((....))))))).))).......)).... (-28.33 = -27.56 + -0.77)


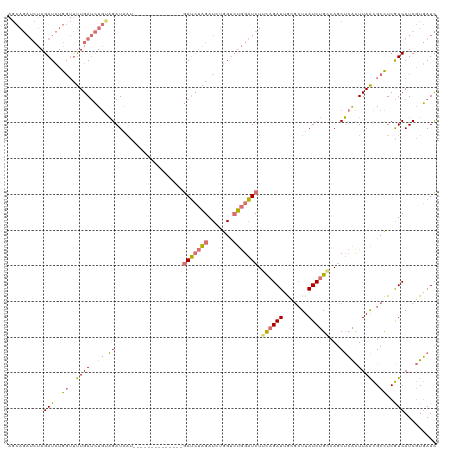
| Location | 6,583,025 – 6,583,145 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.51389 |
| G+C content | 0.43057 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -8.44 |
| Energy contribution | -12.00 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6583025 120 + 27905053 GAUCACGAUGACGAGGCGUUAAGUGAGAUUGCAGAUAAGCCCAAUCUACAGAUCCAGACCAUAACUAGAUCUGCUAUCUCCAAGCAUUAUCUGCAGAUCAAAACCAGAUCUAGAUCUGCU ..((((..(((((...))))).))))((((((((((((((........((((((.((.......)).))))))..........)).))))))))).))).....(((((....))))).. ( -32.47, z-score = -2.46, R) >droSim1.chr3R 12739245 106 - 27517382 GAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCGCAAUCUGAAGAUCCAGACCAGAUCUAGAUC--------------AUUAUCUACAGAUCAAAACCAGAUCUAGAUCUGCU ..((((..(((((...))))).))))(((........)))....((((......)))).((((((((((((--------------.....................)))))))))))).. ( -29.30, z-score = -2.03, R) >droSec1.super_0 5761772 106 - 21120651 GAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCGCAAUCUGAAGAUCCAGACCAGAUCUAGAUC--------------AUUAUCUACAGAUCAAAACCAGAUCUAGAUCUGCU ..((((..(((((...))))).))))(((........)))....((((......)))).((((((((((((--------------.....................)))))))))))).. ( -29.30, z-score = -2.03, R) >droAna3.scaffold_13340 1229188 92 + 23697760 AAUAAGGAUGAUGAAGCACUAAACUGGCUGGCUGAUAACACUUAUCUGUAGAGCUGUGCUUCAUCCCUCUU----------------CAUCC-UACUUCAG---UAUAUCUU-------- ....(((((((((((((((......((((.((.((((.....)))).))..)))))))))))).......)----------------)))))-).......---........-------- ( -25.51, z-score = -2.73, R) >consensus GAUCACGAUGACGAGGCGUUAAGUGAGCUGCCAGAUAAGCCCAAUCUGAAGAUCCAGACCAGAUCUAGAUC______________AUUAUCUACAGAUCAAAACCAGAUCUAGAUCUGCU ..((((..(((((...))))).)))).....(((((.......)))))...........((((((((((((...................................)))))))))))).. ( -8.44 = -12.00 + 3.56)

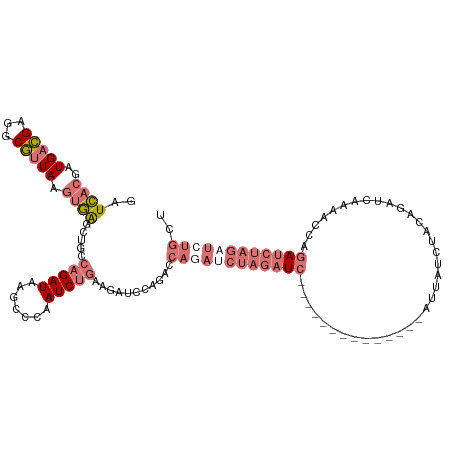

| Location | 6,583,025 – 6,583,145 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.51389 |
| G+C content | 0.43057 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6583025 120 - 27905053 AGCAGAUCUAGAUCUGGUUUUGAUCUGCAGAUAAUGCUUGGAGAUAGCAGAUCUAGUUAUGGUCUGGAUCUGUAGAUUGGGCUUAUCUGCAAUCUCACUUAACGCCUCGUCAUCGUGAUC ..(((((....))))).....(((.(((((((((.(((..(.....((((((((((.......))))))))))...)..)))))))))))))))((((...(((...)))....)))).. ( -39.80, z-score = -1.99, R) >droSim1.chr3R 12739245 106 + 27517382 AGCAGAUCUAGAUCUGGUUUUGAUCUGUAGAUAAU--------------GAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGGCAGCUCACUUAACGCCUCGUCAUCGUGAUC .(.(((((((((((.(((((.((((..........--------------)))).))))).))))))))))).).(((..((.......(((............))).......))..))) ( -33.34, z-score = -1.59, R) >droSec1.super_0 5761772 106 + 21120651 AGCAGAUCUAGAUCUGGUUUUGAUCUGUAGAUAAU--------------GAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGGCAGCUCACUUAACGCCUCGUCAUCGUGAUC .(.(((((((((((.(((((.((((..........--------------)))).))))).))))))))))).).(((..((.......(((............))).......))..))) ( -33.34, z-score = -1.59, R) >droAna3.scaffold_13340 1229188 92 - 23697760 --------AAGAUAUA---CUGAAGUA-GGAUG----------------AAGAGGGAUGAAGCACAGCUCUACAGAUAAGUGUUAUCAGCCAGCCAGUUUAGUGCUUCAUCAUCCUUAUU --------........---.......(-(((((----------------(.((((.((.((((...(((.....((((.....))))....)))..)))).)).)))).))))))).... ( -21.00, z-score = -0.56, R) >consensus AGCAGAUCUAGAUCUGGUUUUGAUCUGUAGAUAAU______________GAUCUAGAUCUGGUCUGGAUCUUCAGAUUGCGCUUAUCUGCCAGCUCACUUAACGCCUCGUCAUCGUGAUC ..........(((..(((.((((..........................(((((((.......))))))).((((((.......))))))........)))).)))..)))......... (-14.52 = -14.40 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:13 2011