| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,571,529 – 6,571,636 |
| Length | 107 |
| Max. P | 0.982862 |

| Location | 6,571,529 – 6,571,636 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.65484 |
| G+C content | 0.45543 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -15.96 |
| Energy contribution | -17.03 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
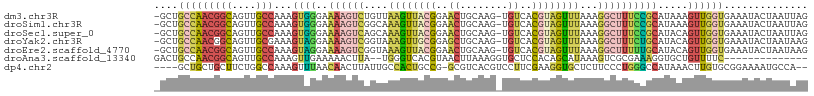
>dm3.chr3R 6571529 107 + 27905053 -GCUGCCAACGGCAGUUGCCAAAGUGGGAAAAGUCUGUUAAGUUACGGAACUGCAAG-UGUCACGUAGUUUAAAGGCUUUCCGCAUAAAGUUGGUGAAAUACUAAUUAG -...(((((((((....)))...((((((..(((((.((((..((((..((......-.))..))))..))))))))))))))).....)))))).............. ( -34.70, z-score = -3.02, R) >droSim1.chr3R 12727618 107 - 27517382 -GCUGCCAACGGCAGUUGCCAAAGUGGGAAAAGUCGGCAAAGUUACGGAACUGCAAG-UGUCACGUAGUUUAAAGGCUUUCCGCAUAAAGUUGGUGAAAUACUAAUUAG -...(((((((((....)))...((((((..((((....((..((((..((......-.))..))))..))...)))))))))).....)))))).............. ( -31.00, z-score = -1.63, R) >droSec1.super_0 5750213 107 - 21120651 -GCUGCCAACGGCAGUUGCCAAAGUGGGAAAAGUCAGCAAAGUUACGGAACUGCAAG-UGUCACGUAGUUUAAAGGCUUUCCGCAUACAGUUGGUGAAAUACUAAUUAG -...(((((((((....)))...((((((..((((....((..((((..((......-.))..))))..))...)))))))))).....)))))).............. ( -29.90, z-score = -1.26, R) >droYak2.chr3R 10611931 107 + 28832112 -GCUGCCAACGGCAGUUGCGAAAGUAGGAAAAGUCGGUAAAGUUGCGGAGCUGCAAG-UGUCACGUAGUUUAAAGGCUUUCUGCAUACAGUUGGUGAAAUACUAAUAAG -...((((((.((((((((....))))..((((((..((((.(((((..((......-.))..)))))))))..)))))))))).....)))))).............. ( -31.60, z-score = -1.67, R) >droEre2.scaffold_4770 2714218 107 - 17746568 -GCUGCCAACGGCAGUUGCCAAAGUAGGAAAAGUCGGUAAAGUUACGGAACUGCAAG-UGUCACGUAGUUUAAAGGCUUUUUGCAUACAGUUGGUGAAAUACUAAUAAG -...(((((((((....)))...(((.((((((((..((((.(((((..((......-.))..)))))))))..))))))))...))).)))))).............. ( -32.10, z-score = -2.43, R) >droAna3.scaffold_13340 1217787 93 + 23697760 GACUGCCAACGGCAGUUGCCAAAGUUGAAAAACUUA--UGGGUCACGUAACUUAAAGGUGCUCCACAGCAUAAAGUCGCGAAAGGUGCUGUUUUC-------------- (((((((...))))))).((((((((....))))).--)))....((..((((....(((((....))))).))))..))...............-------------- ( -26.90, z-score = -1.50, R) >dp4.chr2 1413101 102 - 30794189 ----GCUGCUGCUUCUGGCCAAAGUUUAACAACUUAUUGCCACUGCCG-GCGUCACGUCCUUCGAAGGUGCUCUUCCCUGGGCCAUAAACUUGUGCGGAAAAUGCCA-- ----...((...(((((..(((.(((((..........(((......)-)).....((((...((((.....))))...))))..))))))))..)))))...))..-- ( -23.70, z-score = 1.22, R) >consensus _GCUGCCAACGGCAGUUGCCAAAGUGGGAAAAGUCAGUAAAGUUACGGAACUGCAAG_UGUCACGUAGUUUAAAGGCUUUCCGCAUACAGUUGGUGAAAUACUAAUAAG ....(((((((((....)))...((((..((((((....((((((((..((........))..))))))))...)))))))))).....)))))).............. (-15.96 = -17.03 + 1.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:09 2011