| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,571,051 – 6,571,141 |
| Length | 90 |
| Max. P | 0.610748 |

| Location | 6,571,051 – 6,571,141 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 57.67 |
| Shannon entropy | 0.77535 |
| G+C content | 0.49401 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -7.76 |
| Energy contribution | -7.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.610748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
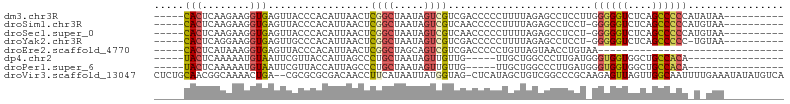
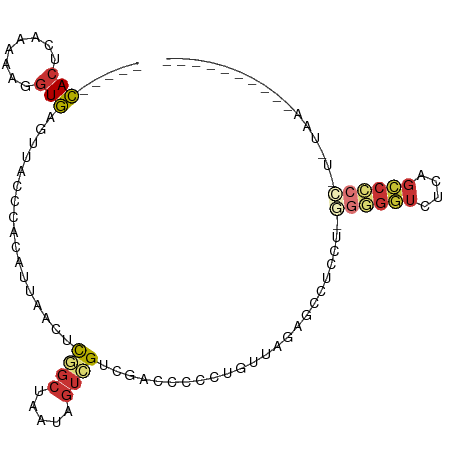
>dm3.chr3R 6571051 90 + 27905053 -----CACUCAAGAAGGUGAGUUACCCACAUUAACUCGGCUAAUAGUCGUCGACCCCCUUUUAGAGCCUCCUUGGGGGUCUCAGCCCCCAUAUAA---------- -----..(((.((((((.((((((.......))))))(((.....)))........)))))).)))......(((((((....))))))).....---------- ( -28.50, z-score = -2.60, R) >droSim1.chr3R 12727141 89 - 27517382 -----CACUCAAGAAGGUGAGUUACCCACAUUAACUCGGCUAAUAGUCGUCAACCCCCUUUUAGAGCCUCCU-GGGGGUCUCAGCCCCCAUGUAA---------- -----..(((.((((((.((((((.......))))))(((.....)))........)))))).))).....(-((((((....))))))).....---------- ( -28.40, z-score = -2.58, R) >droSec1.super_0 5749736 89 - 21120651 -----CACUCAAGAAGGUGAGUUACCCACAUUAACUCGGCUAAUAGUCGUCAACCCCCUUUUAGAGCCUCCU-GGGGGUCUCAGCCCCCAUGUAA---------- -----..(((.((((((.((((((.......))))))(((.....)))........)))))).))).....(-((((((....))))))).....---------- ( -28.40, z-score = -2.58, R) >droYak2.chr3R 10611440 88 + 28832112 -----CACUCAGGAAGGUGAGUUGCCCACAUUAACUCGGCUAAUAGUCGUCGACCCCCUUUUAGAGCCUCCU-GGGGGUCUCAGCCCCC-UGUAA---------- -----....((((..(((((((((.......))))))(((.....)))...(((((((..............-)))))))...))).))-))...---------- ( -28.34, z-score = -1.06, R) >droEre2.scaffold_4770 2713733 69 - 17746568 -----CACUCAUAAAGGUGAGUUACCCACAUUAACUCGGCUAGCAGUCGUCGACCCCCUGUUAGUAACCUGUAA------------------------------- -----.........((((((((((.......)))))).((((((((..(.....)..)))))))).))))....------------------------------- ( -17.80, z-score = -2.28, R) >dp4.chr2 1412754 79 - 30794189 -----UACUCAAAAAUGUAAUUCGUUACCAUUAGCCCUGCUAAUAGUUGUUG-----UUGCUGGCCCUUGAUGGGUGGUGGCUGCCACA---------------- -----(((........)))........(((((((((..((.(((((...)))-----)))).)))...))))))(((((....))))).---------------- ( -18.80, z-score = 0.35, R) >droPer1.super_6 1389050 79 - 6141320 -----UACUCAAAAAUGUAAUUCGUUACCAUUAGCCCUGCUAAUAGUUGUUG-----UUGCUGGCCCUUGAUGGGUGGUGGCUGCCACA---------------- -----(((........)))........(((((((((..((.(((((...)))-----)))).)))...))))))(((((....))))).---------------- ( -18.80, z-score = 0.35, R) >droVir3.scaffold_13047 12093225 102 - 19223366 CUCUGCAACGGCAAAACUGA--CGCGCGCGACAACCUUCAUAAUUAUGGUAG-CUCAUAGCUGUCGGCCCGCAAGAGUUAGUUGGCAAUUUUGAAAUAUAUGUCA ...(((....))).((((((--(..((.((((.(((...........)))((-(.....))))))))).(....).)))))))((((.............)))). ( -23.02, z-score = 0.69, R) >consensus _____CACUCAAAAAGGUGAGUUACCCACAUUAACUCGGCUAAUAGUCGUCGACCCCCUGUUAGAGCCUCCU_GGGGGUCUCAGCCCCC_U_UAA__________ .....(((........))).................((((.....))))........................((((((....))))))................ ( -7.76 = -7.96 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:08 2011