| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,566,484 – 6,566,594 |
| Length | 110 |
| Max. P | 0.887478 |

| Location | 6,566,484 – 6,566,594 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.54448 |
| G+C content | 0.47391 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -15.48 |
| Energy contribution | -17.04 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
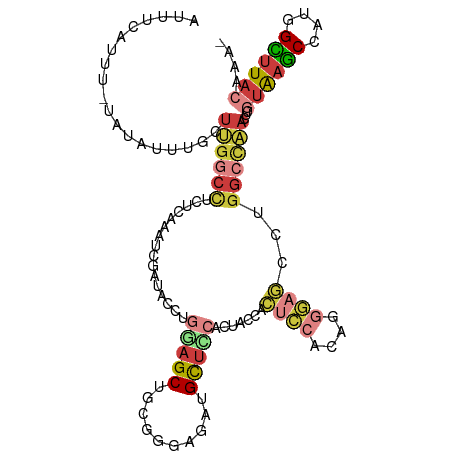
>dm3.chr3R 6566484 110 + 27905053 AAUUCGUUU-AAUAUUUGCUUGGCCUCUCAAAUCGAUACCUGGAGCUGCGGGAGAUGCUCCACUACCACUCCACAGGGAGCCUGGCCAACUCGUAAGCCAAGGCUUACAAA- .........-.........((((((...............((((((..(....)..))))))......((((....))))...))))))...((((((....))))))...- ( -33.50, z-score = -1.62, R) >droSim1.chr3R 12722570 110 - 27517382 AUUUCAUUU-UAUAUUUGCUUGGCCUCUCAAAUCGAUACCUGGAGCUGCGGGAGAUGCUACACUACCACUCCACAGGGAGCCUGGCCAACUCGUAAGCCAAGGCUUACAAA- .........-.........((((((......(((....((((......)))).)))............((((....))))...))))))...((((((....))))))...- ( -28.00, z-score = -0.42, R) >droSec1.super_0 5745206 110 - 21120651 AUUUCAUUU-UAUAUUAGCUUGGCCUCUCAAAUCGAUACCUGGAGCUGCGGGAGAUGCUACACUACCACUCCACAGGGAGCCUGGCCAACUCGUAAGCCAAGGCUUACAAA- .........-......((.((((((......(((....((((......)))).)))............((((....))))...)))))))).((((((....))))))...- ( -28.20, z-score = -0.43, R) >droYak2.chr3R 10606610 110 + 28832112 AUUUCAUUU-UAUAUUUGUUUGGCCUCUCAAAUCGAUACCUGGAGCUGCGGGAGGUGCUCCUACUCCACUCCACAGGGAGCCUGGCCAACUCGUAAGCCAUGGUUUACAAA- .........-.........((((((...............(((((....(((((...))))).)))))((((....))))...))))))...((((((....))))))...- ( -31.30, z-score = -0.86, R) >droEre2.scaffold_4770 2708795 110 - 17746568 AUUUCACUU-UAUAUUUGUUUGGCCUCUCAAAUCGAUACCUGAAGCUGCGGGAAAUGCUUCACUACCACUCCACAGGGUGGCUGGCCAACUCGUAAGCCAUGGCUUACAAG- .........-.........((((((...............((((((..........))))))...((((((....))))))..))))))...((((((....))))))...- ( -33.00, z-score = -2.54, R) >droAna3.scaffold_13340 1212706 103 + 23697760 AUUUCAUUUAUUUAUUUGUUCGCCUCCUCAAAUCGAUAC-UAGAGCAGCGGGA--UGCACGUACCCCACU-----GGGAUCCUGGCUAACUCUUAAGCUAUGGCUUACAAA- ..............(((((..((((((.....((.....-..)).(((.(((.--((....)).))).))-----))))...(((((........))))).)))..)))))- ( -22.00, z-score = -0.36, R) >droMoj3.scaffold_6540 31116554 106 - 34148556 ---UCGAUG-GAAAUAUGCUCAGAGUC-CACAGCGGGCGAUUGAGCUGCGGGUGGUGUUUUGUGUGUGGAA-GUAUCUACGCGAAAAGUGUCUUGAACUUUCGUCUUUAAAA ---..((((-(((....((((((.(((-(.....))))..)))))).......((..((((.((((((((.-...)))))))).))))..))......)))))))....... ( -32.60, z-score = -1.72, R) >consensus AUUUCAUUU_UAUAUUUGCUUGGCCUCUCAAAUCGAUACCUGGAGCUGCGGGAGAUGCUCCACUACCACUCCACAGGGAGCCUGGCCAACUCGUAAGCCAUGGCUUACAAA_ ...................((((((................(((((..........))))).......((((....))))...))))))...((((((....)))))).... (-15.48 = -17.04 + 1.56)

| Location | 6,566,484 – 6,566,594 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.54448 |
| G+C content | 0.47391 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -16.00 |
| Energy contribution | -17.41 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
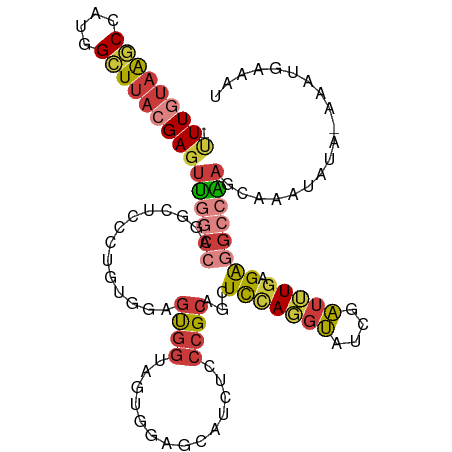
>dm3.chr3R 6566484 110 - 27905053 -UUUGUAAGCCUUGGCUUACGAGUUGGCCAGGCUCCCUGUGGAGUGGUAGUGGAGCAUCUCCCGCAGCUCCAGGUAUCGAUUUGAGAGGCCAAGCAAAUAUU-AAACGAAUU -(((((((((....)))))))))((((((..(((((....)))))((((.((((((..........))))))..)))).........)))))).........-......... ( -38.00, z-score = -1.79, R) >droSim1.chr3R 12722570 110 + 27517382 -UUUGUAAGCCUUGGCUUACGAGUUGGCCAGGCUCCCUGUGGAGUGGUAGUGUAGCAUCUCCCGCAGCUCCAGGUAUCGAUUUGAGAGGCCAAGCAAAUAUA-AAAUGAAAU -(((((((((....)))))))))((((((...(((((((..(.((((.((........)).))))..)..)))).........))).)))))).........-......... ( -33.30, z-score = -0.97, R) >droSec1.super_0 5745206 110 + 21120651 -UUUGUAAGCCUUGGCUUACGAGUUGGCCAGGCUCCCUGUGGAGUGGUAGUGUAGCAUCUCCCGCAGCUCCAGGUAUCGAUUUGAGAGGCCAAGCUAAUAUA-AAAUGAAAU -.((((((((....))))))))((((((..(((...((((((.(.(((.(.....)))).)))))))((((((((....))))).))))))..))))))...-......... ( -36.60, z-score = -1.80, R) >droYak2.chr3R 10606610 110 - 28832112 -UUUGUAAACCAUGGCUUACGAGUUGGCCAGGCUCCCUGUGGAGUGGAGUAGGAGCACCUCCCGCAGCUCCAGGUAUCGAUUUGAGAGGCCAAACAAAUAUA-AAAUGAAAU -((((((.....((((((.((((((((((..(((((....)))))(((((.((((...))))....))))).)))..)))))))..))))))......))))-))....... ( -33.80, z-score = -1.53, R) >droEre2.scaffold_4770 2708795 110 + 17746568 -CUUGUAAGCCAUGGCUUACGAGUUGGCCAGCCACCCUGUGGAGUGGUAGUGAAGCAUUUCCCGCAGCUUCAGGUAUCGAUUUGAGAGGCCAAACAAAUAUA-AAGUGAAAU -(((((((((....)))))))))((((((.(((((((...)).)))))..((((((..........))))))...............)))))).........-......... ( -37.50, z-score = -3.10, R) >droAna3.scaffold_13340 1212706 103 - 23697760 -UUUGUAAGCCAUAGCUUAAGAGUUAGCCAGGAUCCC-----AGUGGGGUACGUGCA--UCCCGCUGCUCUA-GUAUCGAUUUGAGGAGGCGAACAAAUAAAUAAAUGAAAU -....(((((....)))))...(((.(((.......(-----((((((((......)--))))))))(((.(-((....))).)))..))).)))................. ( -26.60, z-score = -1.02, R) >droMoj3.scaffold_6540 31116554 106 + 34148556 UUUUAAAGACGAAAGUUCAAGACACUUUUCGCGUAGAUAC-UUCCACACACAAAACACCACCCGCAGCUCAAUCGCCCGCUGUG-GACUCUGAGCAUAUUUC-CAUCGA--- ..............(((((((.........(.((.((...-.)).)).)............(((((((..........))))))-).)).))))).......-......--- ( -18.20, z-score = -0.81, R) >consensus _UUUGUAAGCCAUGGCUUACGAGUUGGCCAGGCUCCCUGUGGAGUGGUAGUGGAGCAUCUCCCGCAGCUCCAGGUAUCGAUUUGAGAGGCCAAGCAAAUAUA_AAAUGAAAU .(((((((((....)))))))))((((((..............((((..............))))..(((.((((....))))))).))))))................... (-16.00 = -17.41 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:08 2011