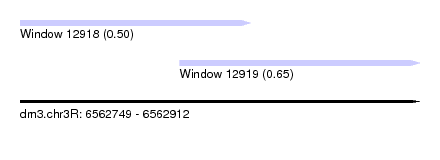
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,562,749 – 6,562,912 |
| Length | 163 |
| Max. P | 0.647873 |

| Location | 6,562,749 – 6,562,843 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.75 |
| Shannon entropy | 0.57686 |
| G+C content | 0.38161 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -9.44 |
| Energy contribution | -8.76 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
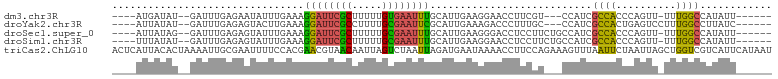
>dm3.chr3R 6562749 94 + 27905053 ----AUGAUAU--GAUUUGAGAAUAUUUGAAAGGAUUCGCUUUUUGUGAAUUUGCAUUGAAGGAACCUUCGU---CCAUCGCCACCCAGUU-UUUGGCCAUAUU------ ----..(((((--(......((.....((...((((((((.....)))))))).))..((((....))))..---...))((((.......-..))))))))))------ ( -19.50, z-score = -0.64, R) >droYak2.chr3R 10602819 95 + 28832112 ----AUUAUAU--GAUUUGAGAGUACUUGAAAGGAUUCGCCUUUUGCGAAUUCGCAUUGAAAGACCCUUUGC---CCAUCGCCACUGAGUCCUUUGGCCUUAUC------ ----.......--..((..((....))..)).((((((((.....))))))))(((..(.......)..)))---.....((((..(....)..))))......------ ( -19.10, z-score = -0.75, R) >droSec1.super_0 5741507 97 - 21120651 ----AUUAUAG--GAUUUGAGAGUAUUUGAAAGGAUUCGCUUUUUGCGAAUUUGCAUUGAAGGGACCUCCUUCUGCCAUCGCCACCCAGUU-UUUGGCCAUAUU------ ----.......--.....((..(((..((...((((((((.....)))))))).))..(((((.....))))))))..))((((.......-..))))......------ ( -21.30, z-score = -0.10, R) >droSim1.chr3R 12718853 97 - 27517382 ----UUUAUAU--GAUUUGAGAGUAUUUGAAAGGAUUCGCUUUUUGCGAAUUUGCAUUGAAGGAACCUCCUUCUGCCAUCGCCACCCAGUU-UUUGGCCAUAUU------ ----...((((--(....((..(((..((...((((((((.....)))))))).))..(((((.....))))))))..))((((.......-..))))))))).------ ( -24.40, z-score = -2.15, R) >triCas2.ChLG10 3448381 110 + 8806720 ACUCAUUACACUAAAAUUGCGAAUUUUCCACGAACGUAACAAUUAGUCUAAUUAGAUGAAUAAAACCUUCCAGAAAGUUUAAUUCUAAUUAGCUGGUCGUCAUUCAUAAU .((.((((.(((((..(((((.............)))))...))))).)))).))((((((...((...((((..((((.......))))..))))..)).))))))... ( -16.82, z-score = -1.51, R) >consensus ____AUUAUAU__GAUUUGAGAGUAUUUGAAAGGAUUCGCUUUUUGCGAAUUUGCAUUGAAGGAACCUUCGU___CCAUCGCCACCCAGUU_UUUGGCCAUAUU______ ................................((((((((.....))))))))...........................((((..........))))............ ( -9.44 = -8.76 + -0.68)

| Location | 6,562,814 – 6,562,912 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Shannon entropy | 0.15916 |
| G+C content | 0.44292 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6562814 98 + 27905053 ---UCCAUCGCCACCCAGU-UUUUGGCCAUAUUUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGGCCAACUGUGUUGAAUGGAAUUGGACUC ---(((((..(.((.((((-...(((((((......(((.....)))..........(((((....)))))))))))))))).)).).)))))......... ( -25.80, z-score = -1.68, R) >droEre2.scaffold_4770 2705093 97 - 17746568 ---CCCAUCGCCACCCAGUCUUUUGGCCAUAUUUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGGCCAACUGGCUUGAAUAGGAUUGGAC-- ---.(((...((..(.((((..((((((((......(((.....)))..........(((((....)))))))))))))..)))).)....))..)))..-- ( -27.10, z-score = -2.09, R) >droYak2.chr3R 10602884 97 + 28832112 ---CCCAUCGCCACUGAGUCCUUUGGCCUUAUCUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGACCAAGUGGGUUGAAUGGAAUUGGAC-- ---.(((...(((.(.(..(((((((..........(((.....)))..........(((((....)))))....))))).))..).).)))...)))..-- ( -21.90, z-score = -0.01, R) >droSec1.super_0 5741572 99 - 21120651 UCUGCCAUCGCCACCCAGU-UUUUGGCCAUAUUUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGGCCAACUGGGUUGAAUGGAAUUGGAC-- ....(((...(((((((((-...(((((((......(((.....)))..........(((((....))))))))))))))))))).....))...)))..-- ( -31.80, z-score = -2.68, R) >droSim1.chr3R 12718918 99 - 27517382 UCUGCCAUCGCCACCCAGU-UUUUGGCCAUAUUUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGGCUAACUGGGUUGAAUGGAUUUGGAC-- ....(((...(((((((((-...(((((((......(((.....)))..........(((((....))))))))))))))))))).....))...)))..-- ( -29.50, z-score = -2.18, R) >consensus ___CCCAUCGCCACCCAGU_UUUUGGCCAUAUUUGAGUAAUAACUACACCCACAUUAGUAUGAAUUCGUGCAUGGCCAACUGGGUUGAAUGGAAUUGGAC__ ....(((...(((((((((....(((((((......(((.....)))..........(((((....))))))))))))))))))).....))...))).... (-22.28 = -23.40 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:04:05 2011