| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,918,572 – 6,918,685 |
| Length | 113 |
| Max. P | 0.660400 |

| Location | 6,918,572 – 6,918,685 |
|---|---|
| Length | 113 |
| Sequences | 10 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 64.66 |
| Shannon entropy | 0.71304 |
| G+C content | 0.43024 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -8.67 |
| Energy contribution | -8.14 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
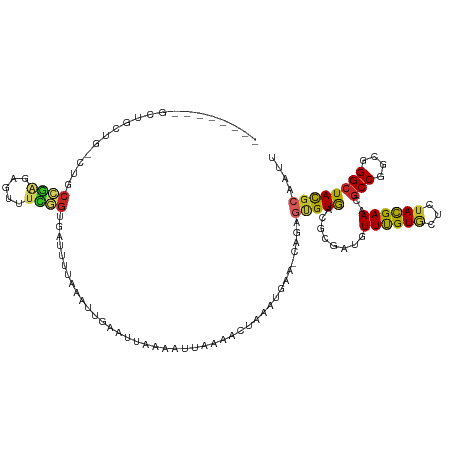
>dm3.chr2L 6918572 113 - 23011544 --------GCUGCUGGCUGCCGAGAGUUUCGGUGGUUUUAAAUUGAAUUAAAAUUGAAACUAAAUGAG-CAGAGCGUAACGCGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCGAUU --------..((((((((((((.(.((((((.(((((((((.((......)).)))))))))...(((-((((((((......)))))).)))))..)))))).))))).))))).)))... ( -40.80, z-score = -3.00, R) >droSim1.chr2L 6714646 113 - 22036055 --------GCUGCUGGCUGCCGAGAGUUUCGGUGGUUUUAAAUUGAAUUAAAAUUGAAACUAAAUGAA-CAGAGUGUGACGCGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCGAUU --------..((((((((((((.(.((((((.(((((((((.((......)).))))))))).(..((-((..(((...)))..))))..)......)))))).))))).))))).)))... ( -37.00, z-score = -2.60, R) >droSec1.super_3 2452304 113 - 7220098 --------GCUGCUGACUGCCGAGAGUUUCGGUGGUUUUAAAUUGAAUUAAAAUUGAAACUAAAUGAA-CAGAGUGUGACGCGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCGAUU --------((.((...((((((.(.((((((.(((((((((.((......)).))))))))).(..((-((..(((...)))..))))..)......)))))).)))))))))...)).... ( -34.10, z-score = -2.16, R) >droYak2.chr2L 16333439 113 + 22324452 --------GCUGCUGGCUGCCGAGAAUUUCGGUGGUUUUAAAUUGAAUUAAAAUUGAAACUAAAUGAG-CAGAGUGUGACGCGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCAAUU --------..((((((((((((.(..(((((.(((((((((.((......)).)))))))))...(((-(((((..(......)..))).)))))..)))))..))))).))))).)))... ( -35.60, z-score = -2.12, R) >droEre2.scaffold_4929 15832240 113 - 26641161 --------GCUGCUGACUGCCGAGAGUUUCGGUGGUUUUAAAUUGAAUUAAAAUUGAAACUAAAUGAG-CAGAGUGUGACGUGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCGAUU --------((.((...((((((.(.((((((.(((((((((.((......)).)))))))))...(((-(((((..(......)..))).)))))..)))))).)))))))))...)).... ( -34.80, z-score = -2.28, R) >droWil1.scaffold_180772 6765424 94 - 8906247 -----------------------UUUCUUUUACCACUGGGGAUUUUGUUUAAAAUAAAA--GAAUUAA-UA-AGUGUGA-GCCAUGUUUGUGCUCUACGAAACGCCUGCGGGCUACGCCAUU -----------------------.((((((((((....))...(((.....))))))))--)))....-..-.((((((-((((....)).))))........(((....))).)))).... ( -17.70, z-score = 0.53, R) >droMoj3.scaffold_6500 14661310 98 + 32352404 -------------------CGUGUCUGCUGCGUGAC-UAAAAUAAUUUUUAACUUAAA--UUAAUGCA-CUAAAAGUG-CAAUAUGUUUGUAUUAUAUGAAACGCCGGCGGGCUACGCAAUU -------------------(((((((((((.(((..-.....((((((.......)))--))).((((-(.....)))-)).....................))))))))))).)))..... ( -25.20, z-score = -2.13, R) >droGri2.scaffold_15126 4611827 102 - 8399593 -------------------CGUGUUUGCUGCGCGACACAAAGUAUUUCUUCACAUAAAUAUUGUUGCAUUUAAGUGUG-CACAAUGUUUGUGUUAUAUGAAACGCCGGCGGGCUACGCAAUU -------------------(((((((((((.(((((.....)).((((...(((((((((((((.((((......)))-)))))))))))))).....))))))))))))))).)))..... ( -35.10, z-score = -2.84, R) >droVir3.scaffold_12963 5454432 99 + 20206255 -------------------CGUGUUUGUUGCGAGACAAAAAAUAGUU-UUGACUUAAA--UUGAUGCAUUUUAACGUG-CACAAUGUUUGUGUUAUAUGAAACGCCGGCGGGCUACGCAAUU -------------------(((((((((((((((((........)))-)))......(--(((.(((((......)))-))))))....(((((......))))))))))))).)))..... ( -21.90, z-score = 0.37, R) >anoGam1.chr2R 45890617 119 + 62725911 GUCGCCGUCUGGCUGGAAUCGGUAAAUUUUAGUGA---AAAAUAAGCUGAUAAGUAUUGGCGUAAGCCUAAAAGUGUAGCACCAUGUUCGUUCUUUACGAAACACCGGCAGGAUAUGCCAUA ...((.((((.(((((....(((..(((((.....---)))))..((((.........(((....)))........))))))).(((((((.....))).))))))))).))))..)).... ( -31.23, z-score = -0.94, R) >consensus ________GCUGCUG_CUGCCGAGAGUUUCGGUGAUUUUAAAUUGAAUUAAAAUUAAAACUAAAUGAA_CAGAGUGUGACGCGAUGUUUGUGCUCUACGAAACGCCGGCGGGCUACGCAAUU ...................((((.....)))).........................................(((((........((((((...))))))..(((....)))))))).... ( -8.67 = -8.14 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:31 2011