| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,550,871 – 6,551,062 |
| Length | 191 |
| Max. P | 0.939541 |

| Location | 6,550,871 – 6,550,962 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.67 |
| Shannon entropy | 0.62823 |
| G+C content | 0.44528 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
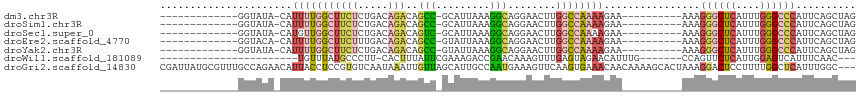
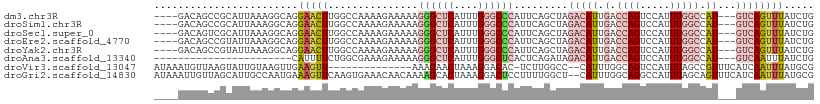
>dm3.chr3R 6550871 91 + 27905053 -------------GGUAUA-CAUUUUGGCUUCUCUGACAGACAGCC-GCAUUAAAGGCAGGAACUUGGCCAAAAGAA----------AAAGGGCUCAUUUGGGCCCAUUCAGCUAG -------------(((...-..(((((((((((.....)))..(((-........)))........))))))))...----------...((((((....)))))).....))).. ( -26.20, z-score = -1.14, R) >droSim1.chr3R 12706735 91 - 27517382 -------------GGUAUA-CAUUUUGGCUUCUCUGACAGACAGCC-GCAUUAAAGGCAGGAACUUGGCCAAAAGAA----------AAAGGGCUCAUUUGGGCCCAUUCAGCUAG -------------(((...-..(((((((((((.....)))..(((-........)))........))))))))...----------...((((((....)))))).....))).. ( -26.20, z-score = -1.14, R) >droSec1.super_0 5729580 91 - 21120651 -------------GGUAUA-CAUGUUGGCUUCUCUGACAGACAGUC-GCAUUAAAGGCAGGAACUUGGCCAAAAGAA----------AAAGGGCUCAUUUGGGCCCAUUCAGCUAG -------------(((...-....((((((..((((((.....)))-((.......)).)))....)))))).....----------...((((((....)))))).....))).. ( -24.50, z-score = -0.65, R) >droEre2.scaffold_4770 2692993 91 - 17746568 -------------GGUACA-CAUUUUGGCUUCUUUGACAGACAGCC-GUAUUAAAGGCAGGAACUUGGCCAAAAGAA----------AAAGGGCUCAUUUGGGCCCAUUCAGCUAG -------------(((...-..(((((((((((.....)))..(((-........)))........))))))))...----------...((((((....)))))).....))).. ( -26.20, z-score = -1.44, R) >droYak2.chr3R 10589556 91 + 28832112 -------------GGUAUA-CAUUUUGGCUUCUCUGACAGACAGCC-GUAUUAAAGGCAGGAACUUGGCCAAAAGAA----------AAAGGGCUCAUUUGGGCCCAUUCAGCUAG -------------(((...-..(((((((((((.....)))..(((-........)))........))))))))...----------...((((((....)))))).....))).. ( -26.20, z-score = -1.31, R) >droWil1.scaffold_181089 7908409 82 - 12369635 -----------------------UGUUUAUGCCCUU-CACUUUAUUCGAAAGACCGAACAAAGUUUGAGUAGAACAUUUG-------CCAGUUCUCAUUGGAGUCAUUUCAAC--- -----------------------((...(((..(((-((((((.((((......)))).)))))....(.(((((.....-------...))))))...)))).)))..))..--- ( -13.90, z-score = -0.35, R) >droGri2.scaffold_14830 3238411 113 - 6267026 CGAUUAUGCGUUUGCCAGAACAUUACCUCCGUGUCAAUAAAUUGUUAGCAUUGCCAAUGAAAGUUCAAGUGAAACAACAAAAGCACUAAAGGACUCCUUUUGGCUCAUUUGGC--- .............((((((...........(((..(((.....)))..))).(((((....(((((.((((............))))...)))))....)))))...))))))--- ( -20.90, z-score = 0.48, R) >consensus _____________GGUAUA_CAUUUUGGCUUCUCUGACAGACAGCC_GCAUUAAAGGCAGGAACUUGGCCAAAAGAA__________AAAGGGCUCAUUUGGGCCCAUUCAGCUAG ......................((((((((..((((.(.................).)))).....))))))))................((((((....)))))).......... (-11.16 = -11.70 + 0.54)
| Location | 6,550,896 – 6,550,999 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.68 |
| Shannon entropy | 0.65101 |
| G+C content | 0.43791 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -10.00 |
| Energy contribution | -11.01 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
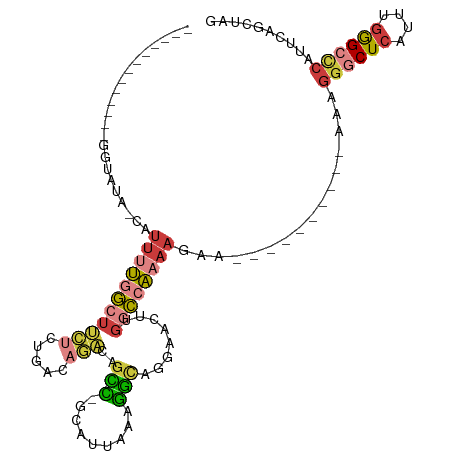
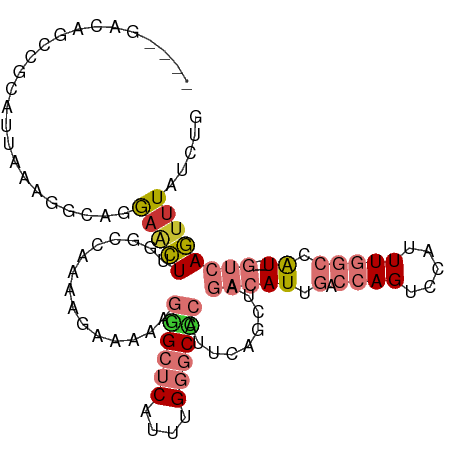
>dm3.chr3R 6550896 103 + 27905053 ----GACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAGUUUAUCUG ----(((((((........))).......((((((((.......((((((....)))))).........(((........)))..)))))))))---))).......... ( -35.10, z-score = -2.72, R) >droSim1.chr3R 12706760 103 - 27517382 ----GACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAGUUUAUCUG ----(((((((........))).......((((((((.......((((((....)))))).........(((........)))..)))))))))---))).......... ( -35.10, z-score = -2.72, R) >droSec1.super_0 5729605 103 - 21120651 ----GACAGUCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAGUUUAUCUG ----(((((((........))).......((((((((.......((((((....)))))).........(((........)))..)))))))))---))).......... ( -32.40, z-score = -2.15, R) >droEre2.scaffold_4770 2693018 103 - 17746568 ----GACAGCCGUAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAGUUUAUCUG ----(((((((........))).......((((((((.......((((((....)))))).........(((........)))..)))))))))---))).......... ( -35.10, z-score = -2.90, R) >droYak2.chr3R 10589581 103 + 28832112 ----GACAGCCGUAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAGUUUAUCUG ----(((((((........))).......((((((((.......((((((....)))))).........(((........)))..)))))))))---))).......... ( -35.10, z-score = -2.90, R) >droAna3.scaffold_13340 1196372 84 + 23697760 -----------------------CAUUUUCUGGCGAAAGAAAAAGGGCUCAUUUGGGCUCACUCAGAUAGACAUUGACCAGUCCAUUUGGCCAU---GUCAAUUUAUCUG -----------------------..((((((......)))))).((((((....))))))...(((((((((((.(.((((.....))))).))---)))....)))))) ( -25.30, z-score = -2.55, R) >droVir3.scaffold_13047 12061978 93 - 19223366 AUAAAUGUUAAGUAUUGUAAGUUGAAGUU--------------AAAGAACUAAAGGACAC-UCUUGGCC--CAUUUGGCAGUCCAUUUAGCCGUUUCAUCAAUUUAUGCG ................(((((((((.(..--------------......(((((((((.(-....)(((--.....))).)))).)))))......).)))))))))... ( -17.64, z-score = -0.75, R) >droGri2.scaffold_14830 3238447 108 - 6267026 AUAAAUUGUUAGCAUUGCCAAUGAAAGUUCAAGUGAAACAACAAAAGCACUAAAGGACUCCUUUUGGCU--CAUUUGGCAGGCCAUUUAGCAGUUUCAUCAAUUUAUGCG ((((((((..(((.((((((((((........((......))....((...(((((...)))))..)))--)).)))))))((......)).)))....))))))))... ( -23.10, z-score = -0.70, R) >consensus ____GACAGCCGCAUUAAAGGCAGGAACUUGGCCAAAAGAAAAAGGGCUCAUUUGGGCCCAUUCAGCUAGACAUUGACCAGUCCAUUUGGCCAU___GUCAGUUUAUCUG ........................(((((...............((((((....)))))).........(((..((.((((.....)))).))....))))))))..... (-10.00 = -11.01 + 1.02)
| Location | 6,550,960 – 6,551,062 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.70 |
| Shannon entropy | 0.31870 |
| G+C content | 0.46441 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
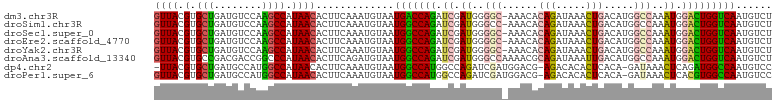
>dm3.chr3R 6550960 102 - 27905053 GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGACCAGAUCGAUGGGGC-AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((..(((-.....(((.....))).....)))..)).)))))))))...... ( -28.50, z-score = -1.66, R) >droSim1.chr3R 12706824 102 + 27517382 GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGGCCAGAUCGAUGGGCC-AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((.((((-(....(((.....)))...)))))..)).)))))))))...... ( -28.80, z-score = -1.48, R) >droSec1.super_0 5729669 102 + 21120651 GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGGCCAGAUCGAUGGGGC-AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((..(((-.....(((.....))).....)))..)).)))))))))...... ( -27.90, z-score = -1.08, R) >droEre2.scaffold_4770 2693082 102 + 17746568 GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGGCCAGAUCGAUGGGGC-AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((..(((-.....(((.....))).....)))..)).)))))))))...... ( -27.90, z-score = -1.08, R) >droYak2.chr3R 10589645 102 - 28832112 GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGGCCAGAUCGAUGGGGC-AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((..(((-.....(((.....))).....)))..)).)))))))))...... ( -27.90, z-score = -1.08, R) >droAna3.scaffold_13340 1196417 103 - 23697760 GUUACGUGCCGACGACCGGCCCAUAACACUUCAGAUGUAAUGGCCAGAUCGAUGGGCCAAAACGCAGAUAAAUUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.((((.....)))).).)))).....((((((..(((((((.((.((.(((((.....(((.....)))...)))))..)).))))))))))))))) ( -32.60, z-score = -1.91, R) >dp4.chr2 1390882 100 + 30794189 -UUACGUGCUGAUGCCAUGGCCAUAACACUUCAAAUGUAAUGGCCAUGGCCAGAUCGAUGGACG-AGACACACUCACA-GAUAAACUCAGAUGGCCAAUGUCC -...((..(((..(((((((((((.(((.......))).))))))))))))))..))..(((((-((.....)))...-((.....))...........)))) ( -32.50, z-score = -3.00, R) >droPer1.super_6 1366995 101 + 6141320 GUUACGUGCUGAUGCCAUGGCCAUAACACUUCAAAUGUAAUGGCCAUGGCCAGAUCGAUGGACG-AGACACACUCACA-GAUAAACUCACGUGGCCAAUGUCC (((((((((((..(((((((((((.(((.......))).))))))))))))))..........(-((.....)))...-........))))))))........ ( -36.30, z-score = -3.58, R) >consensus GUUACGUGCUGAUGUCCAAGCCAUAACACUUCAAAUGUAAUGGCCAGAUCGAUGGGGC_AAACACAGAUAAACUGACAUGGCCAAAUGGACUGGUCAAUGUCU ((((.(.(((........)))).)))).............(((((((.((.((..(((......(((.....))).....)))..)).)))))))))...... (-18.90 = -18.70 + -0.20)
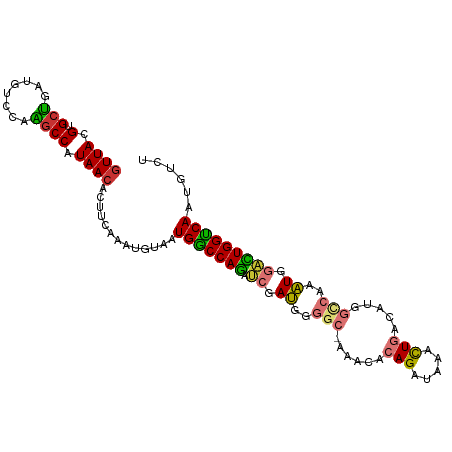
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:59 2011