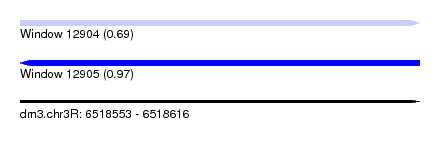
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,518,553 – 6,518,616 |
| Length | 63 |
| Max. P | 0.966660 |

| Location | 6,518,553 – 6,518,616 |
|---|---|
| Length | 63 |
| Sequences | 10 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Shannon entropy | 0.57420 |
| G+C content | 0.64704 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.42 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.690124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
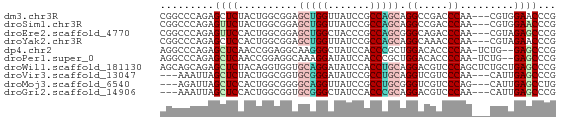
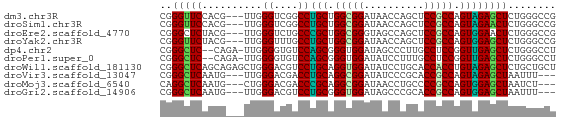
>dm3.chr3R 6518553 63 + 27905053 CGGCCCAGAGCUCUACUGGCGGAGCUGGUUAUCCGCCAGCAGGCCGACCCAA---CGUGGAACCCG (((((.((....)).((((((((........))))))))..)))))..((..---...))...... ( -25.30, z-score = -1.44, R) >droSim1.chr3R 12672152 63 - 27517382 CGGCCCAGAGUUCUACUGGCGGAGCUGGUUAUCCGCCAGCAGGCCGACCCAA---CGUGGAACCCG (((((.((....)).((((((((........))))))))..)))))..((..---...))...... ( -25.30, z-score = -1.68, R) >droEre2.scaffold_4770 2658288 63 - 17746568 CGGCCCAGAGUUCCACUGGCGGAGCUGGCUACCCGCCAGCGGGCAGACCCAA---CGUAGAGCCCG (.(((.((.......))))).).((((((.....))))))((((..((....---.))...)))). ( -24.40, z-score = -0.81, R) >droYak2.chr3R 10556137 63 + 28832112 CGGCCCAGAGCUCCACUGGCGGAGCUGGUUAUCCGCCAGCAGGCAAACCCAA---CGUAGAACCCG .........(((...((((((((........))))))))..)))........---........... ( -18.90, z-score = -0.32, R) >dp4.chr2 23856843 63 - 30794189 AGGCCCAGAGCUCAACCGGAGGCAAGGGCUAUCCACCCGCUGGACACCCCAA-UCUG--GAGCCCG .(((((((((((........)))..(((...((((.....))))...)))..-))))--).))).. ( -23.30, z-score = -1.11, R) >droPer1.super_0 11029485 63 - 11822988 AGGCCCAGAGCUCAACCGGAGGCAAAGGAUAUCCACCCGCUGGACACCCCAA-UCUG--GAGCCCG .((((((((......((((.((....((....))..)).)))).........-))))--).))).. ( -20.06, z-score = -1.07, R) >droWil1.scaffold_181130 8090446 66 - 16660200 AGCAGCAGAGCUCUACAGGUGGUGCAGGAUAUCCACCUGCAGGACGUCCCAGCUCUGCUGAGCCCG ..((((((((((...(((((((..........)))))))..((.....))))))))))))...... ( -30.50, z-score = -2.57, R) >droVir3.scaffold_13047 1514135 60 + 19223366 ---AAAUUAGCUCUACUGGCGGUGCGGGAUAUCCGCCUGCAGGUCGUCCCAA---CAUUGAGCCCG ---......((((...((((((.((((.....)))))))).((.....))..---))..))))... ( -19.30, z-score = -0.91, R) >droMoj3.scaffold_6540 5608219 60 + 34148556 ---AGAUUAGCUCCACUGGCGGGGCAGGUUAUCCGCCUGCGGGUCGUCCCAG---CAUUGAGCCUG ---......((((....((((((........))))))((((((....))).)---))..))))... ( -22.30, z-score = -0.46, R) >droGri2.scaffold_14906 9452257 60 - 14172833 ---AAAUUAGCUCCACUGGCGGUGCGGGCUAUCCACCCGCAGGACGUCCCAA---CAUUGAGCCCG ---......((((....((((.((((((.......))))))...))))....---....))))... ( -18.00, z-score = -0.58, R) >consensus CGGCCCAGAGCUCCACUGGCGGAGCUGGUUAUCCGCCAGCAGGACGACCCAA___CGUUGAGCCCG .........((((..........(((((.......))))).((.....)).........))))... (-11.43 = -11.42 + -0.01)
| Location | 6,518,553 – 6,518,616 |
|---|---|
| Length | 63 |
| Sequences | 10 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 71.68 |
| Shannon entropy | 0.57420 |
| G+C content | 0.64704 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -16.50 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
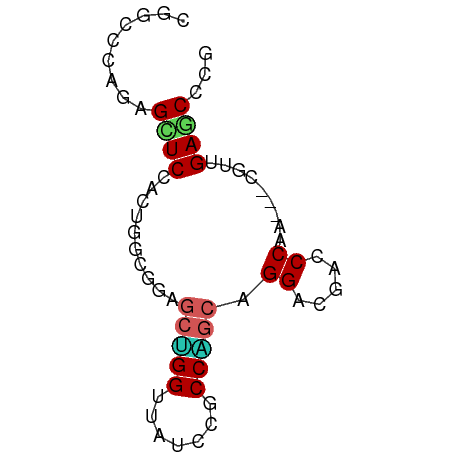
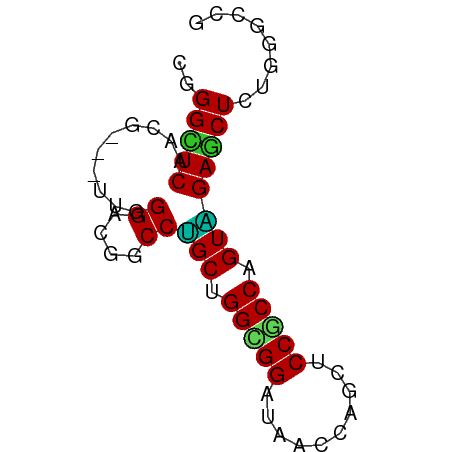
>dm3.chr3R 6518553 63 - 27905053 CGGGUUCCACG---UUGGGUCGGCCUGCUGGCGGAUAACCAGCUCCGCCAGUAGAGCUCUGGGCCG ..(((.(((((---......))(((((((((((((........))))))))))).))..)))))). ( -29.80, z-score = -1.70, R) >droSim1.chr3R 12672152 63 + 27517382 CGGGUUCCACG---UUGGGUCGGCCUGCUGGCGGAUAACCAGCUCCGCCAGUAGAACUCUGGGCCG ..(((.(((..---..((((....(((((((((((........))))))))))).)))))))))). ( -27.60, z-score = -1.42, R) >droEre2.scaffold_4770 2658288 63 + 17746568 CGGGCUCUACG---UUGGGUCUGCCCGCUGGCGGGUAGCCAGCUCCGCCAGUGGAACUCUGGGCCG ..(((((...(---(((((....)))((((((((((.....)).))))))))..)))...))))). ( -31.20, z-score = -1.37, R) >droYak2.chr3R 10556137 63 - 28832112 CGGGUUCUACG---UUGGGUUUGCCUGCUGGCGGAUAACCAGCUCCGCCAGUGGAGCUCUGGGCCG .(((((((((.---..(((....)))..(((((((........))))))))))))))))....... ( -27.60, z-score = -1.44, R) >dp4.chr2 23856843 63 + 30794189 CGGGCUC--CAGA-UUGGGGUGUCCAGCGGGUGGAUAGCCCUUGCCUCCGGUUGAGCUCUGGGCCU .((((.(--((((-..(((.((((((.....)))))).)))..((.((.....))))))))))))) ( -27.70, z-score = -0.36, R) >droPer1.super_0 11029485 63 + 11822988 CGGGCUC--CAGA-UUGGGGUGUCCAGCGGGUGGAUAUCCUUUGCCUCCGGUUGAGCUCUGGGCCU .((((.(--((((-..((((((((((.....))))))))))..((.((.....))))))))))))) ( -27.00, z-score = -0.81, R) >droWil1.scaffold_181130 8090446 66 + 16660200 CGGGCUCAGCAGAGCUGGGACGUCCUGCAGGUGGAUAUCCUGCACCACCUGUAGAGCUCUGCUGCU ......(((((((((((((....)))((((((((..........))))))))..)))))))))).. ( -33.20, z-score = -2.63, R) >droVir3.scaffold_13047 1514135 60 - 19223366 CGGGCUCAAUG---UUGGGACGACCUGCAGGCGGAUAUCCCGCACCGCCAGUAGAGCUAAUUU--- ..(((((..((---(.((.....)).)))(((((..........)))))....))))).....--- ( -21.00, z-score = -1.36, R) >droMoj3.scaffold_6540 5608219 60 - 34148556 CAGGCUCAAUG---CUGGGACGACCCGCAGGCGGAUAACCUGCCCCGCCAGUGGAGCUAAUCU--- ..(((((..((---(.(((....))))))(((((..........)))))....))))).....--- ( -23.70, z-score = -1.59, R) >droGri2.scaffold_14906 9452257 60 + 14172833 CGGGCUCAAUG---UUGGGACGUCCUGCGGGUGGAUAGCCCGCACCGCCAGUGGAGCUAAUUU--- ..(((((.((.---...((.((...(((((((.....))))))).)))).)).))))).....--- ( -23.20, z-score = -0.98, R) >consensus CGGGCUCAACG___UUGGGACGGCCUGCUGGCGGAUAACCAGCUCCGCCAGUAGAGCUCUGGGCCG ..(((((..........((....))(((.(((((..........))))).))))))))........ (-16.50 = -15.75 + -0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:53 2011