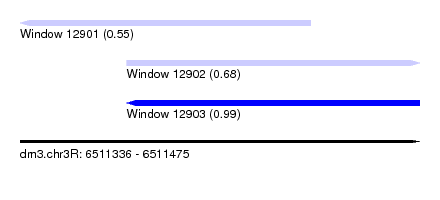
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,511,336 – 6,511,475 |
| Length | 139 |
| Max. P | 0.990861 |

| Location | 6,511,336 – 6,511,437 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.84 |
| Shannon entropy | 0.56354 |
| G+C content | 0.35882 |
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -7.66 |
| Energy contribution | -7.20 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
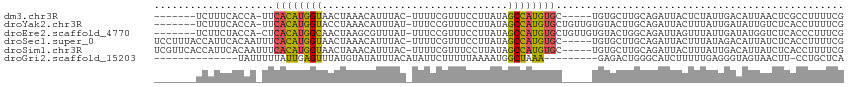
>dm3.chr3R 6511336 101 - 27905053 -------UCUUUCACCA-UUCACAUGGUAACUAAACAUUUAC-UUUUCGUUUCCUUAUAGCCAUGUGC-----UGUGCUUGCAGAUUACUCUAUUGACAUUAACUCGCCUUUUCG -------....(((...-..((((((((((..((((......-.....))))..))...))))))))(-----(((....))))..........))).................. ( -12.70, z-score = -0.86, R) >droYak2.chr3R 10548834 106 - 28832112 -------UCUUUCACCA-UUCACAUGGUACCUAAACAUUUAU-UUUCCGUUUCCUUAUAGCCAUGUGCUGUUGUGUACUUGCAGAUUACUUUAUUGAUAUUGUCUCACCUUUUCG -------.....(((.(-(.((((((((....((((......-.....)))).......))))))))..)).))).....((((((((......)))).))))............ ( -12.70, z-score = -0.46, R) >droEre2.scaffold_4770 2650973 106 + 17746568 -------UCUUCUACCA-CUCACAUGGCAACUAAGCGUUUAU-UUUCCGUUUCCUUAUAGCCAUGUGCUGUUGUGUACUGGCAGAUUAGUUUAUUGAUAUGGUCUCACCCUUUCG -------......((((-..((((((((....(((((.....-....))))).......))))))))(((((.......)))))...............))))............ ( -19.40, z-score = -0.45, R) >droSec1.super_0 5690286 109 + 21120651 UCCUUUACCAUUCACAAUUUCACAUGGUAACUAAACAUUUAC-UUUUCGUUUCCUUAUAGCCAUGUGC-----UGUGCUUGCAGAUUACUUUAUAGACAUUAUCUCACCUUUUCG ........((..((((....((((((((((..((((......-.....))))..))...)))))))).-----))))..)).................................. ( -13.20, z-score = -1.30, R) >droSim1.chr3R 12665010 109 + 27517382 UCGUUCACCAUUCACAAUUUCACAUGGUAACUAAACAUUUAC-UUUUCGUUUCCUUAUAGCCAUGUGC-----UGUGCUUGCAGAUUACUUUAUUGACAUUAUCUCACCUUUUCG ....((..((..((((....((((((((((..((((......-.....))))..))...)))))))).-----))))..))..)).............................. ( -13.50, z-score = -0.94, R) >droGri2.scaffold_15203 448660 91 + 11997470 --------------UAUUUUUAUUGAGUUUAUGUAUAUUUACAUAUUCUUUUUAAAAUGGCUAAA---------GAGACUGGGCAUCUUUUUGAGGGUAGUAACUU-CCUGCUCA --------------...(((((..(((..((((((....))))))..)))..)))))....((((---------((((.......)))))))).((((((......-.)))))). ( -20.40, z-score = -2.00, R) >consensus _______UCUUUCACAA_UUCACAUGGUAACUAAACAUUUAC_UUUUCGUUUCCUUAUAGCCAUGUGC_____UGUGCUUGCAGAUUACUUUAUUGACAUUAUCUCACCUUUUCG ....................((((((((...............................))))))))................................................ ( -7.66 = -7.20 + -0.47)
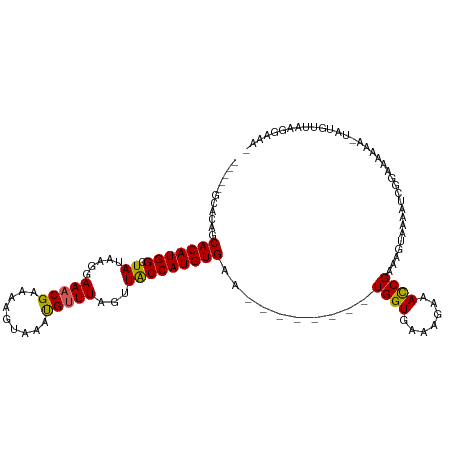
| Location | 6,511,373 – 6,511,475 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.29214 |
| G+C content | 0.35743 |
| Mean single sequence MFE | -20.04 |
| Consensus MFE | -12.20 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
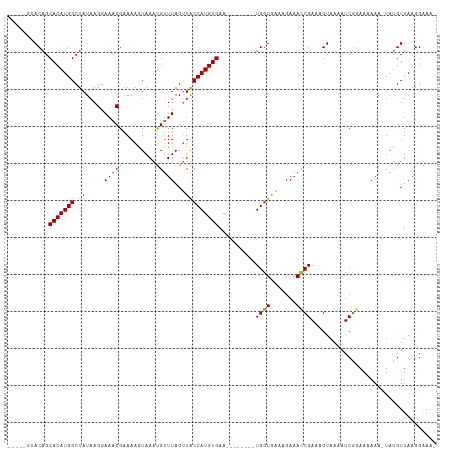
>dm3.chr3R 6511373 102 + 27905053 -----GCACAGCACAUGGCUAUAAGGAAACGAAAAGUAAAUGUUUAGUUACCAUGUGAA--------UGGUGAAAGAAACCGAAAGUAAAAUCAGAAAAAAAUAUGUUAAGGAAA- -----.......((((((((....(....)....)))....((((..(((((((....)--------))))))...))))(....)................)))))........- ( -15.60, z-score = -1.37, R) >droYak2.chr3R 10548871 107 + 28832112 GUACACAACAGCACAUGGCUAUAAGGAAACGGAAAAUAAAUGUUUAGGUACCAUGUGAA--------UGGUGAAAGAAACCGAGAGUAAGAUCGCAAGAAA-UGUGUUAAGGGAAC ..(((((((..(((((((.(((....(((((.........)))))..))))))))))..--------((((.......))))...))....((....))..-)))))......... ( -23.00, z-score = -2.71, R) >droEre2.scaffold_4770 2651010 107 - 17746568 GUACACAACAGCACAUGGCUAUAAGGAAACGGAAAAUAAACGCUUAGUUGCCAUGUGAG--------UGGUAGAAGAAAUCGAAAGUAAAAUCGAAAAAAA-UGUGUUAAGGGAAG ..(((((.((.((((((((.....(....)........(((.....)))))))))))..--------))..........((((........))))......-)))))......... ( -24.00, z-score = -3.26, R) >droSec1.super_0 5690323 109 - 21120651 -----GCACAGCACAUGGCUAUAAGGAAACGAAAAGUAAAUGUUUAGUUACCAUGUGAAAUUGUGAAUGGUAAAGGAAACCGAAAGUAAAAUCGGAAAAAA-UACGUUAAGGAAG- -----.((((.(((((((......(....)...((.(((....))).)).)))))))....))))..............((((........))))......-.............- ( -18.80, z-score = -1.59, R) >droSim1.chr3R 12665047 109 - 27517382 -----GCACAGCACAUGGCUAUAAGGAAACGAAAAGUAAAUGUUUAGUUACCAUGUGAAAUUGUGAAUGGUGAACGAAACCGAAAGUAAAAUCGGAAAAAA-UAUGUUAAGGAAG- -----.((((.(((((((......(....)...((.(((....))).)).)))))))....))))..............((((........))))......-.............- ( -18.80, z-score = -1.42, R) >consensus _____GCACAGCACAUGGCUAUAAGGAAACGAAAAGUAAAUGUUUAGUUACCAUGUGAA________UGGUGAAAGAAACCGAAAGUAAAAUCGGAAAAAA_UAUGUUAAGGAAA_ ...........(((((((.((.....(((((.........)))))...)))))))))..........((((.......)))).................................. (-12.20 = -11.92 + -0.28)

| Location | 6,511,373 – 6,511,475 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.29214 |
| G+C content | 0.35743 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.24 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
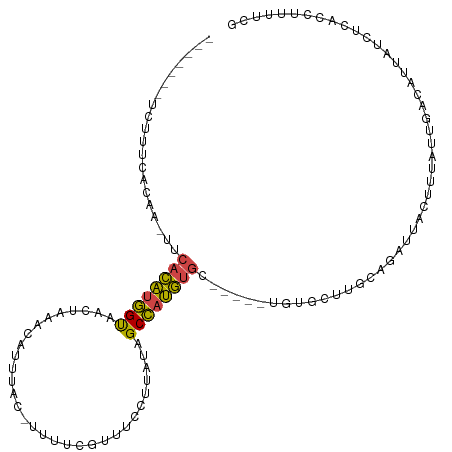
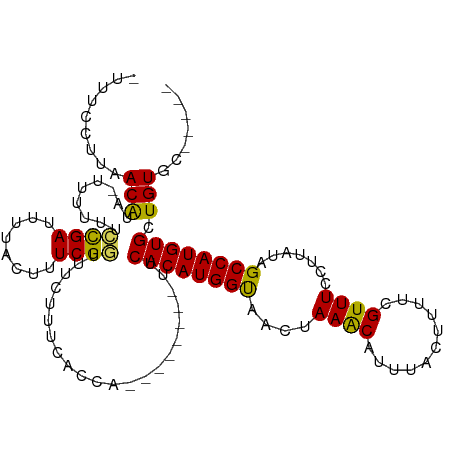
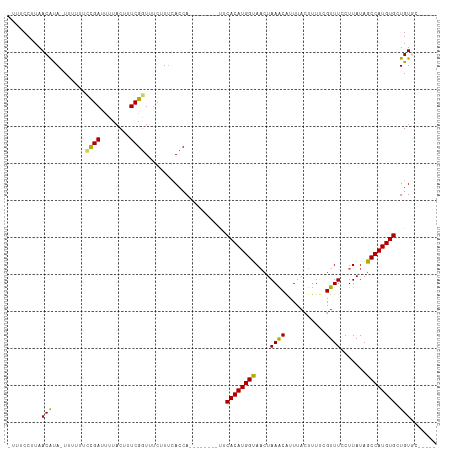
>dm3.chr3R 6511373 102 - 27905053 -UUUCCUUAACAUAUUUUUUUCUGAUUUUACUUUCGGUUUCUUUCACCA--------UUCACAUGGUAACUAAACAUUUACUUUUCGUUUCCUUAUAGCCAUGUGCUGUGC----- -....................((((........))))..........((--------(.((((((((((..((((...........))))..))...))))))))..))).----- ( -13.00, z-score = -1.77, R) >droYak2.chr3R 10548871 107 - 28832112 GUUCCCUUAACACA-UUUCUUGCGAUCUUACUCUCGGUUUCUUUCACCA--------UUCACAUGGUACCUAAACAUUUAUUUUCCGUUUCCUUAUAGCCAUGUGCUGUUGUGUAC .........(((((-....................(((.......))).--------..((((((((....((((...........)))).......))))))))....))))).. ( -15.80, z-score = -1.79, R) >droEre2.scaffold_4770 2651010 107 + 17746568 CUUCCCUUAACACA-UUUUUUUCGAUUUUACUUUCGAUUUCUUCUACCA--------CUCACAUGGCAACUAAGCGUUUAUUUUCCGUUUCCUUAUAGCCAUGUGCUGUUGUGUAC .........(((((-......((((........))))...........(--------(.((((((((....(((((.........))))).......))))))))..))))))).. ( -20.90, z-score = -4.29, R) >droSec1.super_0 5690323 109 + 21120651 -CUUCCUUAACGUA-UUUUUUCCGAUUUUACUUUCGGUUUCCUUUACCAUUCACAAUUUCACAUGGUAACUAAACAUUUACUUUUCGUUUCCUUAUAGCCAUGUGCUGUGC----- -..........(((-......((((........)))).......)))....((((....((((((((((..((((...........))))..))...)))))))).)))).----- ( -17.32, z-score = -3.11, R) >droSim1.chr3R 12665047 109 + 27517382 -CUUCCUUAACAUA-UUUUUUCCGAUUUUACUUUCGGUUUCGUUCACCAUUCACAAUUUCACAUGGUAACUAAACAUUUACUUUUCGUUUCCUUAUAGCCAUGUGCUGUGC----- -.............-......((((........))))..............((((....((((((((((..((((...........))))..))...)))))))).)))).----- ( -16.00, z-score = -2.50, R) >consensus _UUUCCUUAACAUA_UUUUUUCCGAUUUUACUUUCGGUUUCUUUCACCA________UUCACAUGGUAACUAAACAUUUACUUUUCGUUUCCUUAUAGCCAUGUGCUGUGC_____ .........(((.........((((........))))......................((((((((....((((...........)))).......)))))))).)))....... (-12.92 = -12.24 + -0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:52 2011