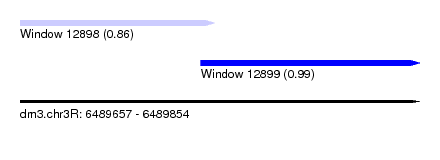
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,489,657 – 6,489,854 |
| Length | 197 |
| Max. P | 0.987629 |

| Location | 6,489,657 – 6,489,753 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.41847 |
| G+C content | 0.61374 |
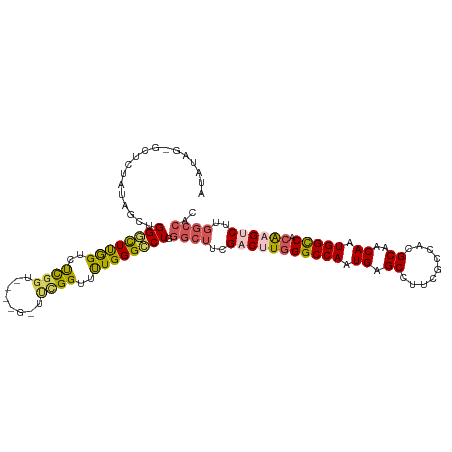
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6489657 96 + 27905053 CGGUGUGCGCGCUCUUUUGAGUUAUGCCACUGGGGU---GAUGGAGCCCCACCCCCCAGA---UCCCGCCCUG-AAAGUGGCCCCCACCAGU--GCACAUAUAGU------------ ..((((((((((((....))))...(((((((((((---(.........)))))).(((.---.......)))-..))))))........))--)))))).....------------ ( -37.70, z-score = -2.84, R) >droSec1.super_0 5667562 96 - 21120651 CGGUGUGCGCGCUCUUUUGAGUUAUGCCACUGGGGU---GAUGGAGCCCCACCCCC-AGA---UCCCGCCCUGCAAAGUGCCCCCCACCAGU--GGACAUAUAGU------------ .((((.((((((((....)))).......((((((.---(.(((....))))))))-)).---..............))))....))))...--...........------------ ( -29.10, z-score = -0.09, R) >droYak2.chr3R 10524618 116 + 28832112 CGGUGUGCGCGCUCUUUUGAGUUAUGCCAGUGGGGCUCCAUCGCAGCCCCACCCCCACAUCCCUCCCUCCCUG-AAAGUGCCCCCCACCCACUGGGGCACAUAGAUAUGUAUAUAGA .((((((.((((((....))))...))..((((((((.......))))))))...))))))............-...(((((((.........)))))))................. ( -39.50, z-score = -1.98, R) >droEre2.scaffold_4770 2627775 84 - 17746568 CGGUGUGCGCGCUCUUUUGAGUUAUGCCACUGGGGCUCCAUCGCAGCCCCACCCCAGA-----UCCCUUCCUG-AAACUGCCCCCCACCA--------------------------- .((((((.((((((....))))...)))))(((((((.......)))))))...(((.-----.......)))-............))).--------------------------- ( -24.40, z-score = -1.42, R) >consensus CGGUGUGCGCGCUCUUUUGAGUUAUGCCACUGGGGC___AACGCAGCCCCACCCCC_AGA___UCCCGCCCUG_AAAGUGCCCCCCACCAGU__GGACAUAUAGU____________ .((((.((((((((....))))........(((((((.......)))))))..........................))))....))))............................ (-21.20 = -21.95 + 0.75)

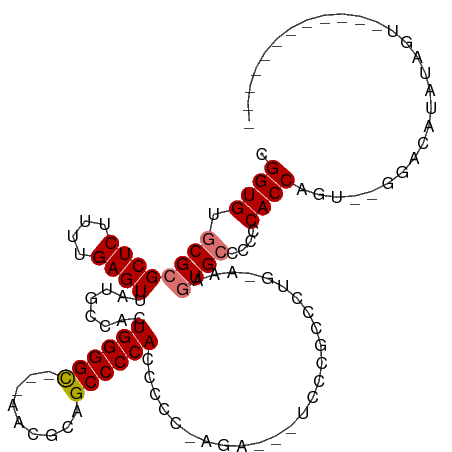
| Location | 6,489,746 – 6,489,854 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.80 |
| Shannon entropy | 0.50744 |
| G+C content | 0.57656 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6489746 108 + 27905053 AUAUAGUGCUCUAUAGCUGGGCUUGGUCUCGGU------UUCGGUUUUGGGCCUGGGCUUCGACUUGGGCCAAUGAGCCUUCGCCACGCAACAAUGGCCACAAGACUUGGCCAC .(((((....)))))....((((.(((((.(..------...(((((..((((..((......))..))))...)))))...((((........))))..).))))).)))).. ( -39.70, z-score = -1.35, R) >droSec1.super_0 5667651 108 - 21120651 AUAUAGUGCUCUAUAGCUGGGCUUGGUCUCGGU------UUCGGUUUUGGGCCUGGGCUUCGACUUGGGCCAAUGAGCCUUCGCCACGCAACAAUGGCCACAAGUCUUGGCCAC .(((((....)))))...(((((..(..(((..------..)))..)..))))).((((..(((((((((((.((.((.........))..)).))))).))))))..)))).. ( -41.00, z-score = -1.66, R) >droYak2.chr3R 10524727 114 + 28832112 AUAUAGAGCUGUAUAGUUGGGCUUGGUCUCGGUCUCGGUUUCGGUUUUGGGCCUGGGCUUCGACUUGGGCCAAUGAGCCUUCGCCACGCAACAAUGGCCACAAGUCUUGGCCAC (((((....)))))....(((((..(..((((........))))..)..))))).((((..(((((((((((.((.((.........))..)).))))).))))))..)))).. ( -40.70, z-score = -0.91, R) >droEre2.scaffold_4770 2627859 105 - 17746568 --------AGCUGGGCUUGGUCUCGGUUUCGGUUUUGGCUAUGGGCCUGGGCCUGGGCUUCGACUUGGGCCAAUGGGCCUUCGCCACGCAACAAUGGCCACA-GUCUUGGCCAC --------.((..(((((((((...(((.((....((((...((((((.((((..((......))..))))...))))))..)))))).)))...))))).)-).))..))... ( -48.00, z-score = -2.12, R) >droWil1.scaffold_181108 302698 95 + 4707319 ------------------GGUUUUAGGCCUGGGA-CAGACCGGGGACUGGGACUGGGCCUGGGCCUAGGCCAAUGAGCCUUCGCCAUGCAACAAUGGUCAGGCGACAUUAAACC ------------------(((((.(((((..((.-(((.(((....).))..)))..))..))))))))))((((.((((..(((((......))))).))))..))))..... ( -44.00, z-score = -3.28, R) >consensus AUAUAG_GCUCUAUAGCUGGGCUUGGUCUCGGU____G_UUCGGUUUUGGGCCUGGGCUUCGACUUGGGCCAAUGAGCCUUCGCCACGCAACAAUGGCCACAAGUCUUGGCCAC ..................((((((((..((((........))))..)))))))).((((..(((((((((((.((.((.........))..)).))))).))))))..)))).. (-28.32 = -28.88 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:49 2011