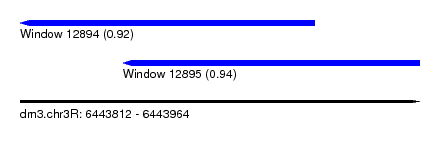
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,443,812 – 6,443,964 |
| Length | 152 |
| Max. P | 0.943940 |

| Location | 6,443,812 – 6,443,924 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.30727 |
| G+C content | 0.40095 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -18.72 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
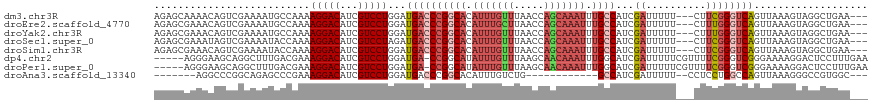
>dm3.chr3R 6443812 112 - 27905053 GAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUUC---UUCGGGUCAGUUAAAGUAGGC---UGAAUAAUGACUGGAGAAAAAUUAAAUAAAGCUCGCAGGACCU ......((((((.(((((((.....))))))).))))...((((((((---(((((.((((((......)))---))).......)))))))))))))..............)).... ( -31.90, z-score = -2.69, R) >droEre2.scaffold_4770 2580562 112 + 17746568 GAUGACCCGGCACAUUUGCUUAACCAGCAAAUUUGCCAUCGAUUUUUC---UUUGGGUCAGUUAAAGUAGGC---UGAAUGAUGACUGGAGGAAAAUCAAACAAAACUCGAAGGACCU ......((((((.(((((((.....))))))).))))...((((((((---((..(.((((((......)))---))).......)..))))))))))..............)).... ( -34.90, z-score = -3.27, R) >droYak2.chr3R 10475995 112 - 28832112 GAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUUC---UUUGGGUCAGUUAAAGUAGGC---UGAACGAUGACUGGAAGAAAAUCAAAUAAAACUCGAAGGACCU ......((((((.(((((((.....))))))).))))...((((((.(---(((.(((((.....((....)---)......))))).))))))))))..............)).... ( -30.40, z-score = -2.33, R) >droSec1.super_0 5622022 112 + 21120651 GAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUUC---UUCGGGUCAGUUAAAGUAGGC---UGAAUAAUGACUGGAGAAAACUUAAAUAAAACUCGCAGGACCC ......((((((.(((((((.....))))))).))))......(((((---(((((.((((((......)))---))).......)))))))))).................)).... ( -28.30, z-score = -1.82, R) >droSim1.chr3R 12591790 112 + 27517382 GAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUUC---UUCGGGUCAGUUAAAGUAGGC---UGAAUAAUGACUGGAGAAAACUUAAAUAAAACUCGCAGGACCC ......((((((.(((((((.....))))))).))))......(((((---(((((.((((((......)))---))).......)))))))))).................)).... ( -28.30, z-score = -1.82, R) >dp4.chr2 23773762 117 + 30794189 GAUGA-CCGGCAUAUUUGUUUAAGCAACAAAUUUGGCAUCGAUUUUUCGUUUUCGGGUCGGGAAAAGGACUCCUUUGAAAGAUGAGUCGAGGAAAAAAAACCAAAAAACGAAAAAUCU ((((.-((((...(((((((.....)))))))))))))))(((((((((((((..(((.........(((((..((....)).)))))...........)))..))))))))))))). ( -34.25, z-score = -2.98, R) >droPer1.super_0 10949383 114 + 11822988 GAUGA-CCGGCAUAUUUGUUUAAGCAACAAAUUUGGCAUCGAUUUUUCGUUUUCGGGUCGGGAAAAGGACUCCUUUGAAAGAUGAGUCGAGGAUAAAAA--CGAAAAACGAAAAAUC- ((((.-((((...(((((((.....)))))))))))))))(((((((((((((((((((........)))))((((((........)))))).......--.).)))))))))))))- ( -32.80, z-score = -2.79, R) >consensus GAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUUC___UUCGGGUCAGUUAAAGUAGGC___UGAAUGAUGACUGGAGGAAAAUUAAAUAAAACUCGAAGGACCU ..((((((((((.(((((((.....))))))).))))...((....))......)))))).......(((...............))).............................. (-18.72 = -17.95 + -0.77)
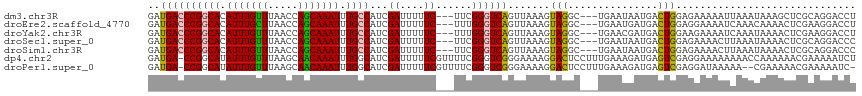
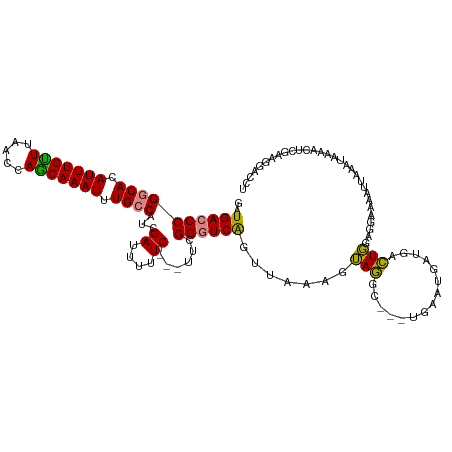
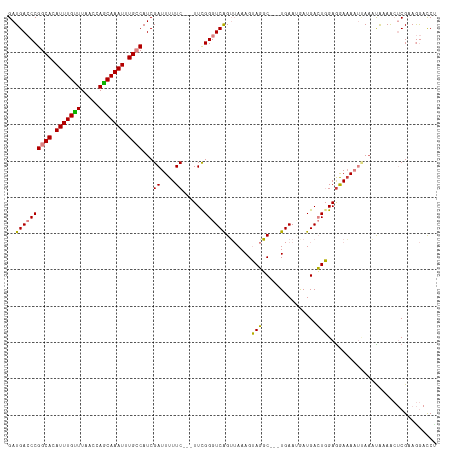
| Location | 6,443,851 – 6,443,964 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Shannon entropy | 0.45680 |
| G+C content | 0.45774 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -18.86 |
| Energy contribution | -20.11 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.943940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
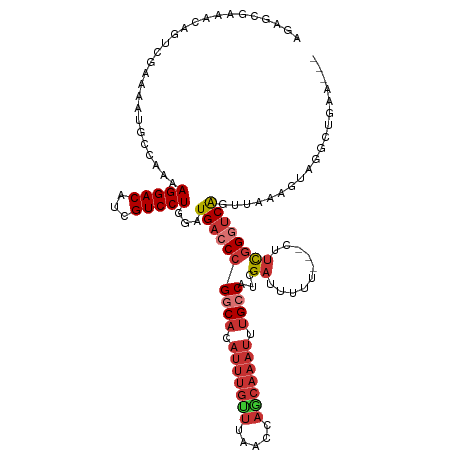
>dm3.chr3R 6443851 113 - 27905053 AGAGCAAAACAGUCGAAAAUGCCAAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUU---CUUCGGGUCAGUUAAAGUAGGCUGAA--- .........(((((.......(((...((((...))))))).((((((((((.(((((((.....))))))).))))...((.....---..)))))))).........)))))..--- ( -34.90, z-score = -2.81, R) >droEre2.scaffold_4770 2580601 113 + 17746568 AGAGCGAAACAGUCGAAAAUGCCAAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGCUUAACCAGCAAAUUUGCCAUCGAUUUUU---CUUUGGGUCAGUUAAAGUAGGCUGAA--- .........(((((.......(((...((((...))))))).((((((((((.(((((((.....))))))).))))...((....)---)...)))))).........)))))..--- ( -36.80, z-score = -2.97, R) >droYak2.chr3R 10476034 113 - 28832112 AGAGCGAAACAGUCGAAAAUGCCAAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUU---CUUUGGGUCAGUUAAAGUAGGCUGAA--- .........(((((.......(((...((((...))))))).((((((((((.(((((((.....))))))).))))...((....)---)...)))))).........)))))..--- ( -34.40, z-score = -2.41, R) >droSec1.super_0 5622061 113 + 21120651 AGAGCGAAAUAGUCGAAAAUACCAAAAGGACAUCGUCCUAGAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUU---CUUCGGGUCAGUUAAAGUAGGCUGAA--- .(((.((((.((((((...........((.((((......)))).)).((((.(((((((.....))))))).)))).)))))))))---))))...((((((......)))))).--- ( -32.50, z-score = -2.95, R) >droSim1.chr3R 12591829 113 + 27517382 AGAGCGAAACAGUCGAAAAUACCAAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUU---CUUCGGGUCAGUUAAAGUAGGCUGAA--- .........(((((.......(((...((((...))))))).((((((((((.(((((((.....))))))).))))...((.....---..)))))))).........)))))..--- ( -35.20, z-score = -3.07, R) >dp4.chr2 23773801 113 + 30794189 -----AGGGAAGCAGGCUUUGACGAAAGGACAUCGUCCUGGAUGA-CCGGCAUAUUUGUUUAAGCAACAAAUUUGGCAUCGAUUUUUCGUUUUCGGGUCGGGAAAAGGACUCCUUUGAA -----...((((....))))....((((((....(((((...(((-(((.((.(((((((.....))))))).)).)..(((....)))......))))).....)))))))))))... ( -30.00, z-score = -0.24, R) >droPer1.super_0 10949419 113 + 11822988 -----AGGGAAGCAGGCUUUGACGAAAGGACAUCGUCCUGGAUGA-CCGGCAUAUUUGUUUAAGCAACAAAUUUGGCAUCGAUUUUUCGUUUUCGGGUCGGGAAAAGGACUCCUUUGAA -----...((((....))))....((((((....(((((...(((-(((.((.(((((((.....))))))).)).)..(((....)))......))))).....)))))))))))... ( -30.00, z-score = -0.24, R) >droAna3.scaffold_13340 23031446 95 + 23697760 -------AGGCCCGGCAGAGCCCGAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGUCUG------------GCCAUCGAUUUUU--CCUCCUGGCCAGUUAAAGGGCCGUGGC--- -------.((((((((((((.((....)).)...((.((((.....)))).)).))))))((------------((((..((....)--)....))))))......))))).....--- ( -37.00, z-score = -1.19, R) >consensus AGAGCGAAACAGUCGAAAAUGCCAAAAGGACAUCGUCCUGGAUGACCCGGCACAUUUGUUUAACCAGCAAAUUUGCCAUCGAUUUUU___CUUCGGGUCAGUUAAAGUAGGCUGAA___ ..........................(((((...)))))...((((((((((.(((((((.....))))))).))))...((..........))))))))................... (-18.86 = -20.11 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:45 2011