| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,886,298 – 6,886,419 |
| Length | 121 |
| Max. P | 0.877962 |

| Location | 6,886,298 – 6,886,419 |
|---|---|
| Length | 121 |
| Sequences | 6 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 74.99 |
| Shannon entropy | 0.44836 |
| G+C content | 0.53391 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.95 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6886298 121 + 23011544 -----CGC-CCCCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGCAAUGAAUAAAAACGCUGACUGCUCCGUCCGUCCGUUGUGCCCGUCCGGCUCUUCAGCC--GGAUGAACGAGUGGUGUGCGUG------ -----.((-......))(((((....))))).....(((((......)))))......((((..((..(((.((.(((((((((.(((.....)))....))).)--))))).)))))..))..)))).------ ( -39.40, z-score = -2.09, R) >droSim1.chr2L 6681818 127 + 22036055 CGCUCCACGCCCCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGCAAUGAAUAAAAACGCUGACUGCUCCGUCCGUUCGUCGUGCCCGUCCGGCUCUUCAGCC--GGAUGAACGAGUGGUGUGCGUG------ .....(((((.(.(..((((.((((((.........(((((......)))))........)))))).)))...........((((...(((((((((....))))--))))).)))))..).).)))))------ ( -43.03, z-score = -2.25, R) >droSec1.super_3 2420193 127 + 7220098 CGCUCCACGCCCCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGCAAUGAAUAAAAACGCUGACUGCUCCGUCCGUCCGUCGUGCCCGUCCGGCUCUUCAGCC--GGAUGAACGAGUGGUGUGCGUG------ .....(((((.(.(..((((.((((((.........(((((......)))))........)))))).)))...........((((...(((((((((....))))--))))).)))))..).).)))))------ ( -43.03, z-score = -2.30, R) >droYak2.chr2L 16300580 129 - 22324452 ---CUCCCCAACCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGAAAUGAAUAAAAACGCUGCCAGCUCCGUCCGUCGUUC---CUCGUCCGGCUCUCUAGUCUAGGAUGAAUGAGUGGUUUGCGUGUGCGUG ---............(((((((....))))).....((((((....))))))......((((.((((.(((((((((.((...---..)).).((((....))))..)))))...)))))))..))))..))... ( -33.30, z-score = -1.14, R) >droEre2.scaffold_4929 15799776 107 + 26641161 --CUCCCCCUUCCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGAAAUGAAUAAAAACGCAGGCUGCUCCGACUC------------------CCCUCUAUCC--GGAUGAAUGAGUGGUGUGCGUG------ --...............(((((....))))).....((((((....))))))......(((((.((..(((....((------------------(.........--))).....)))..)).))))).------ ( -28.50, z-score = -1.91, R) >droAna3.scaffold_12916 8467644 104 + 16180835 CAUCCUGUGGCUGCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUUGAAAUGAAUAAAAACGCUGUCUGU--CGACGUU-----------------CUUUCUAUUC------GUAUGACUGGUGUGCGUG------ .......((((((((((....).)))))))))....((((((....))))))......(((((..(.((--(((((..-----------------.........)------)).)))).)..).)))).------ ( -28.70, z-score = -2.22, R) >consensus __CUCCACGCCCCCUGCGGCUGUCAGUAGCCAAUUAUCAUUUUUCGAAAUGAAUAAAAACGCUGACUGCUCCGUCCGUCCGUC___CCCGUCCGGCUCUCCAGCC__GGAUGAACGAGUGGUGUGCGUG______ .................(((((....))))).....(((((......)))))......((((..((((((((((((...........((....))............)))))...)))))))..))))....... (-18.23 = -19.95 + 1.72)
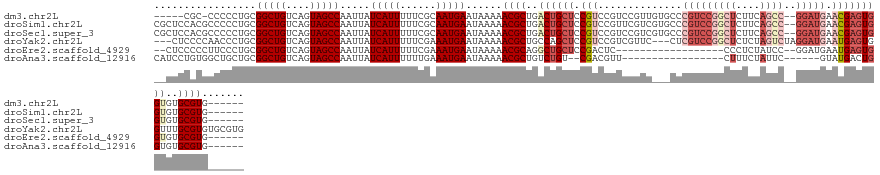


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:29 2011