
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,381,190 – 6,381,308 |
| Length | 118 |
| Max. P | 0.985452 |

| Location | 6,381,190 – 6,381,308 |
|---|---|
| Length | 118 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Shannon entropy | 0.40713 |
| G+C content | 0.39138 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6381190 118 + 27905053 AAAGGAACAAAAGUAUAAGUACGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCCGGGC-GAAAAAGUUGUUUAAAUU ....((((....(((....))).......(((((((.((((.((((..(((.(((((((.(((..((......))..)))..))))))).))))))))))-).)))))))))))..... ( -32.80, z-score = -2.34, R) >droSim1.chr3R 12527301 118 - 27517382 AAAGGAACAAAAGUAUAAGUACGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCCGGGC-GAAAAAGUUGUUUAAAUU ....((((....(((....))).......(((((((.((((.((((..(((.(((((((.(((..((......))..)))..))))))).))))))))))-).)))))))))))..... ( -32.80, z-score = -2.34, R) >droSec1.super_0 5559619 118 - 21120651 AAAGAAACAAAAGUAUAAGUACGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCCCGGC-GAAAAAGUUGUUUAAAUU ....((((....(((....))).......(((((((.((((..((.(((((.(((((((.(((..((......))..)))..))))))).))))))))))-).)))))))))))..... ( -33.30, z-score = -2.84, R) >droYak2.chr3R 10411116 118 + 28832112 AAAGGCACAAGAGUAUAAGUACGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCCGGGC-GAAAAAGUUGUUUAAAUU ...(((((((((((................))))))))))).((.((((((.(((((((.(((..((......))..)))..))))))).)))))..).)-)................. ( -35.19, z-score = -2.65, R) >droEre2.scaffold_4770 2517706 117 - 17746568 AAAGGAACGAGCGCAUAAGUGCGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCC-GGC-GAAAAAGUUGUUUAAAUU ....((((..((((....)))).......(((((((.((((...(.(((((.(((((((.(((..((......))..)))..))))))).))))))-)))-).)))))))))))..... ( -34.70, z-score = -1.74, R) >droAna3.scaffold_13340 22976129 104 - 23697760 ---AAAGCGAAAACGCCAGC-CGAAAUGGAACUUUUGCGCCAAGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCAUUAUAUG----GGC-------GUUGUUUAAAUU ---.(((((...(((((.((-...((((((((((((((((((((......))))))))...)))))......)))))))....))........----)))-------)))))))..... ( -28.30, z-score = -1.05, R) >dp4.chr2 23709027 96 - 30794189 ----------------------GAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGU-ACGGGUACGGGCAGAAAAAGUUGUUUAAAUU ----------------------.......(((((((.((((..((((((....)))))).(((..((......))..)))....(((-(....))))))))..)))))))......... ( -27.10, z-score = -1.76, R) >droPer1.super_0 10884393 96 - 11822988 ----------------------GAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGU-ACGGGUACGGGCAGAAAAAGUUGUUUAAAUU ----------------------.......(((((((.((((..((((((....)))))).(((..((......))..)))....(((-(....))))))))..)))))))......... ( -27.10, z-score = -1.76, R) >droVir3.scaffold_13047 1350913 98 + 19223366 ----------AAAAAACCAAACAAAAUGAAACUUUU--GCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCAUUGCG------GGC---AAAAGUUGUUUAAAUU ----------...................(((((((--(((.(((((((....)))))..(((..((......))..))).........))------)))---)))))))......... ( -26.70, z-score = -2.52, R) >droMoj3.scaffold_6540 5423545 98 + 34148556 ----------AAGAAACCAAACAAAAUGAAACUUUU--GCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCAUUGCA------GGC---AAAAGUUGUUUAAAUU ----------...................(((((((--(((..((((((....)))))).(((..((......))..)))...........------)))---)))))))......... ( -26.60, z-score = -2.81, R) >consensus ___G_AAC_AAAGUAUAAGUACGAAAUGGAACUUUUGUGCCACGGCGCCAUUUGGCGCUACAAAAGAUAUUUUUCCAUUGAAAGCGUUAUAUGGCCGGGC_GAAAAAGUUGUUUAAAUU .............................(((((((..(((..((((((....)))))).(((..((......))..))).................)))...)))))))......... (-21.42 = -21.82 + 0.40)

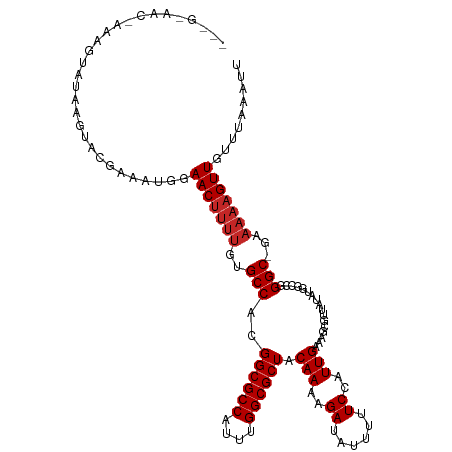
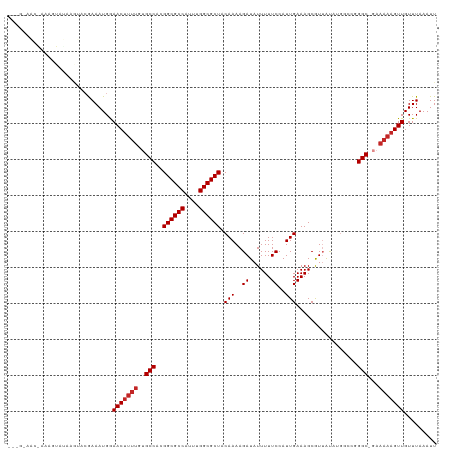
| Location | 6,381,190 – 6,381,308 |
|---|---|
| Length | 118 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Shannon entropy | 0.40713 |
| G+C content | 0.39138 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -17.81 |
| Energy contribution | -18.21 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6381190 118 - 27905053 AAUUUAAACAACUUUUUC-GCCCGGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGUACUUAUACUUUUGUUCCUUU ..................-(((((((........(((..(((.(((......))).))).)))(((....))))))).)))((((((((................))))))))...... ( -29.89, z-score = -3.30, R) >droSim1.chr3R 12527301 118 + 27517382 AAUUUAAACAACUUUUUC-GCCCGGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGUACUUAUACUUUUGUUCCUUU ..................-(((((((........(((..(((.(((......))).))).)))(((....))))))).)))((((((((................))))))))...... ( -29.89, z-score = -3.30, R) >droSec1.super_0 5559619 118 + 21120651 AAUUUAAACAACUUUUUC-GCCGGGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGUACUUAUACUUUUGUUUCUUU ..................-(((((((........(((..(((.(((......))).))).)))(((....)))))).))))((((((((................))))))))...... ( -27.69, z-score = -2.16, R) >droYak2.chr3R 10411116 118 - 28832112 AAUUUAAACAACUUUUUC-GCCCGGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGUACUUAUACUCUUGUGCCUUU .........(((((((..-(((((((........(((..(((.(((......))).))).)))(((....))))))).)))..))))))).......((((..........)))).... ( -30.60, z-score = -3.06, R) >droEre2.scaffold_4770 2517706 117 + 17746568 AAUUUAAACAACUUUUUC-GCC-GGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGCACUUAUGCGCUCGUUCCUUU ......((((((((((..-(((-(((........(((..(((.(((......))).))).)))(((....))))))..)))..)))))))......((((....))))...)))..... ( -26.90, z-score = -1.16, R) >droAna3.scaffold_13340 22976129 104 + 23697760 AAUUUAAACAAC-------GCC----CAUAUAAUGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCUUGGCGCAAAAGUUCCAUUUCG-GCUGGCGUUUUCGCUUU--- .........(((-------(((----........(((...(((((((......(((((((...(((....)))((....))))))))))))))))..)-)).))))))........--- ( -27.90, z-score = -1.29, R) >dp4.chr2 23709027 96 + 30794189 AAUUUAAACAACUUUUUCUGCCCGUACCCGU-ACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUC---------------------- .........(((((((..((((((...((((-.........))))................(((((....))))))).)))).))))))).......---------------------- ( -22.70, z-score = -2.04, R) >droPer1.super_0 10884393 96 + 11822988 AAUUUAAACAACUUUUUCUGCCCGUACCCGU-ACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUC---------------------- .........(((((((..((((((...((((-.........))))................(((((....))))))).)))).))))))).......---------------------- ( -22.70, z-score = -2.04, R) >droVir3.scaffold_13047 1350913 98 - 19223366 AAUUUAAACAACUUUU---GCC------CGCAAUGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGC--AAAAGUUUCAUUUUGUUUGGUUUUUU---------- ...(((((((((((((---(((------(((...(((..(((.(((......))).))).)))(((....)))).)).)))--)))))).......)))))))......---------- ( -27.81, z-score = -2.49, R) >droMoj3.scaffold_6540 5423545 98 - 34148556 AAUUUAAACAACUUUU---GCC------UGCAAUGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGC--AAAAGUUUCAUUUUGUUUGGUUUCUU---------- ...(((((((((((((---(((------.((...(((..(((.(((......))).))).)))(((....)))))...)))--)))))).......)))))))......---------- ( -27.71, z-score = -2.70, R) >consensus AAUUUAAACAACUUUUUC_GCCCGGCCAUAUAACGCUUUCAAUGGAAAAAUAUCUUUUGUAGCGCCAAAUGGCGCCGUGGCACAAAAGUUCCAUUUCGUACUUAUACUUU_GUU_C___ .........(((((((...(((.................(((.(((......))).)))..(((((....)))))...)))..)))))))............................. (-17.81 = -18.21 + 0.40)

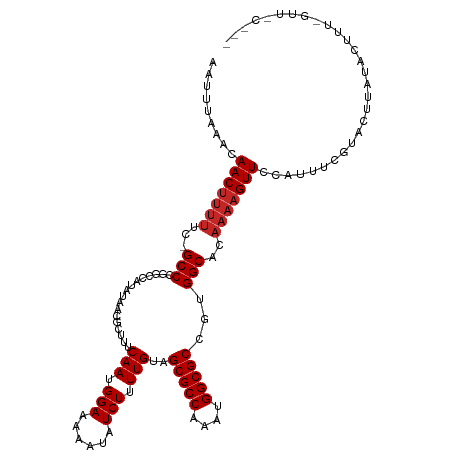
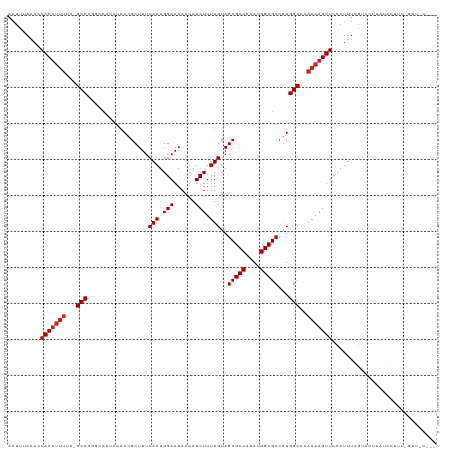
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:37 2011