
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,377,133 – 6,377,231 |
| Length | 98 |
| Max. P | 0.646747 |

| Location | 6,377,133 – 6,377,223 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 86.79 |
| Shannon entropy | 0.26467 |
| G+C content | 0.44361 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
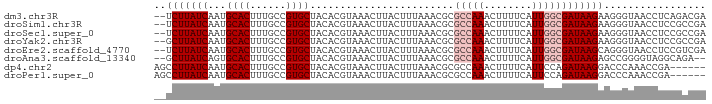
>dm3.chr3R 6377133 90 + 27905053 --UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCAGACGA --(((((((...((((......))))........................(((((........))))))))))))(((....)))....... ( -21.30, z-score = -2.14, R) >droSim1.chr3R 12523370 90 - 27517382 --UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGA --(((((((...((((......))))........................(((((........))))))))))))(((....)))....... ( -21.30, z-score = -1.82, R) >droSec1.super_0 5555682 90 - 21120651 --UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGA --(((((((...((((......))))........................(((((........))))))))))))(((....)))....... ( -21.30, z-score = -1.82, R) >droYak2.chr3R 10406851 90 + 28832112 --GCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGA --.((((((...((((......))))........................(((((........))))))))))).(((....)))....... ( -19.90, z-score = -0.96, R) >droEre2.scaffold_4770 2513792 90 - 17746568 --UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGCAGGGUAACCUCCGUCGA --.....((.(((....(((((.((((....(((....)))........((((((........))))))...)))).)))))....))).)) ( -20.90, z-score = -1.58, R) >droAna3.scaffold_13340 22972088 88 - 23697760 --GCUUAUCAGUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAGCCGGGGUAGGCAGA-- --(((((((.((((((......)))).)).......((..((((.....((((((........))))))...))))..)))))))))...-- ( -25.10, z-score = -1.14, R) >dp4.chr2 23704908 86 - 30794189 AGCCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUCCAGAUAAGGACCCAAACCGA------ ..(((((((...((((......)))).....(((....)))...........................)))))))...........------ ( -14.30, z-score = -1.65, R) >droPer1.super_0 10880240 86 - 11822988 AGCCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUCCAGAUAAGGACCCAAACCGA------ ..(((((((...((((......)))).....(((....)))...........................)))))))...........------ ( -14.30, z-score = -1.65, R) >consensus __UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGA ..(((((((...((((......))))........................(((((........))))))))))))................. (-14.69 = -15.62 + 0.94)


| Location | 6,377,133 – 6,377,231 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.28246 |
| G+C content | 0.46335 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -14.64 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6377133 98 + 27905053 UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCAGACGAAACCCUGC (((((((...((((......))))........................(((((........))))))))))))(((((.............))))).. ( -24.32, z-score = -2.67, R) >droSim1.chr3R 12523370 98 - 27517382 UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGAAUCCCUGC (((((((...((((......))))........................(((((........))))))))))))((((...............)))).. ( -22.96, z-score = -2.02, R) >droSec1.super_0 5555682 98 - 21120651 UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGAAUCCCUGC (((((((...((((......))))........................(((((........))))))))))))((((...............)))).. ( -22.96, z-score = -2.02, R) >droYak2.chr3R 10406851 98 + 28832112 GCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGAGUCCCUGC .((((((...((((......))))........................(((((........))))))))))).((((....(((....))).)))).. ( -24.20, z-score = -1.56, R) >droEre2.scaffold_4770 2513792 98 - 17746568 UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGCAGGGUAACCUCCGUCGAGUCCCUGC ..(((((...((((......))))........................(((((........))))))))))((((((....(((....))).)))))) ( -26.40, z-score = -2.59, R) >droAna3.scaffold_13340 22972088 96 - 23697760 GCUUAUCAGUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGA--GCCGGGGUAGGCAGAGUCCCUGG ......(((.(.(((((((((((...))).((..((..((((.....((((((........))))))...)))--)..))..)))))))))).)))). ( -27.90, z-score = -0.95, R) >dp4.chr2 23704910 97 - 30794189 CCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUCCAGAUAAGGA-CCCAAACCGAAGCGAACCCCAGC (((((((...((((......)))).....(((....)))...........................))))))).-...........((........)) ( -14.30, z-score = -0.99, R) >droPer1.super_0 10880242 87 - 11822988 CCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUCCAGAUAAGGA-CCCAAACCGAACC---------- (((((((...((((......)))).....(((....)))...........................))))))).-.............---------- ( -14.10, z-score = -2.33, R) >consensus UCUUAUCAAUGCACUUUGCCGUGCUACACGUAAACUUACUUUAAACGCGCCAAACUUUUCAUUGGCGAUAAGAAGGGUAACCUCCGCCGAAUCCCUGC (((((((...((((......))))........................(((((........))))))))))))......................... (-14.64 = -15.57 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:35 2011