
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,359,447 – 6,359,571 |
| Length | 124 |
| Max. P | 0.603501 |

| Location | 6,359,447 – 6,359,571 |
|---|---|
| Length | 124 |
| Sequences | 10 |
| Columns | 133 |
| Reading direction | forward |
| Mean pairwise identity | 61.46 |
| Shannon entropy | 0.80001 |
| G+C content | 0.40196 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -7.80 |
| Energy contribution | -8.31 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
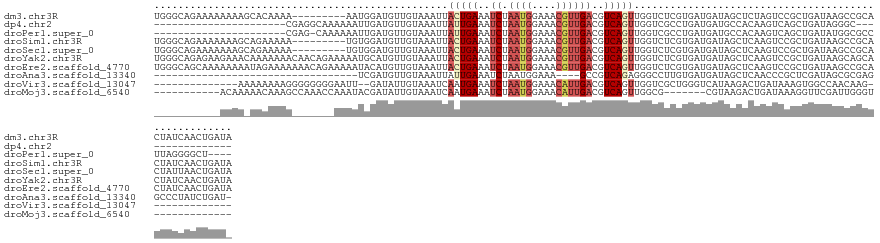
>dm3.chr3R 6359447 124 + 27905053 UGGGCAGAAAAAAAAGCACAAAA---------AAUGGAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCUAGUCCGCUGAUAAGCCGCACUAUCAACUGAUA (((((..................---------...(((((((((.....(((((..((.((((....))))))..)))))..(((....))).)))))....))))(((....))).)).))).......... ( -24.40, z-score = 0.17, R) >dp4.chr2 23687238 95 - 30794189 ----------------------CGAGGCAAAAAAUUGAUGUUGUAAAUUAUUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCGCCUGAUGAUGCCACAAGUCAGCUGAUAGGGC---------------- ----------------------..((((.(..((((((((((..........(....).((((....))))))))))))))..).))))......(((....(((....)))..)))---------------- ( -23.00, z-score = -1.15, R) >droPer1.super_0 10862473 106 - 11822988 ----------------------CGAG-CAAAAAAUUGAUGUUGUAAAUUAUUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCGCCUGAUGAUGCCACAAGUCAGCUGAUAUGGCGCCUUAGGGGCU---- ----------------------....-.....((((((((((..........(....).((((....))))))))))))))((((.(((((.(.(((((...(((....))).)))))).)))))))))---- ( -28.30, z-score = -1.19, R) >droSim1.chr3R 12496277 124 - 27517382 UGGGCAGAAAAAAAGCAGAAAAA---------UGUGGAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCAAGUCCGCUGAUAAGCCGCACUAUCAACUGAUA ...............(((.....---------.(((((((((((.....(((((..((.((((....))))))..)))))..(((....))).)))))....))))))(((((........))))).)))... ( -26.60, z-score = -0.17, R) >droSec1.super_0 5538269 124 - 21120651 UGGGCAGAAAAAAAGCAGAAAAA---------UGUGGAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCAAGUCCGCUGAUAAGCCGCACUAUUAACUGAUA .((((.........(((......---------))).((.(((((.....(((((..((.((((....))))))..)))))..(((....))).)))))))..))))(((....)))................. ( -25.70, z-score = -0.07, R) >droYak2.chr3R 10388285 133 + 28832112 UGGGCAGAGAAGAAACAAAAAAACAACAGAAAAAUGCAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCAAGUCCGCUGAUAAGCAGCACUAUCAACUGAUA .((((.((((............((((((..........)))))).....(((((..((.((((....))))))..)))))...))))....(((....))).))))((((.....))))..((((....)))) ( -32.80, z-score = -1.96, R) >droEre2.scaffold_4770 2496595 133 - 17746568 UGGGCAGCAAAAAAAAUAGAAAAAAACAGAAAAAUACAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCAAGUCCGCUGAUAAGCCGCACUAUCAACUGAUA ....((((........((((......(((.((..(((.....)))..)).)))...))))..(....)))))..((((((((((...(((.(((....))).....(((....))))))...)))))))))). ( -24.90, z-score = -0.27, R) >droAna3.scaffold_13340 22955870 94 - 23697760 ----------------------------------UCGAUGUUGUAAAUUAUUGAAAUCUAAUGGAAA----GCCGUCAGAGGGCCUUGUGAUGAUAGCUCAACCCGCUCGAUAGCGCGAGGCCCUAUCUGAU- ----------------------------------((((((........))))))........((...----.))((((((((((((((((.....(((.......)))......))))))))))..))))))- ( -30.90, z-score = -2.82, R) >droVir3.scaffold_13047 1328405 103 + 19223366 --------------AAAAAAAAGGGGGGGGAAUU--GAUAUUGUAAAUCAAUGAAAUCUAAUGGAAACAUUGACGUCAGUUGGUCGCUGGGUCAUAAGACUGAUAAAGUGGCCAACAAG-------------- --------------.................(((--(((.......))))))((..((.((((....))))))..)).((((((((((..((((......))))..))))))))))...-------------- ( -30.60, z-score = -5.00, R) >droMoj3.scaffold_6540 5399964 102 + 34148556 -----------ACAAAAACAAAGCCAAACCAAAUACGAUAUUGUAAAUCAAUGAAAUCUAAUGGAAACAUUGACGUCAGUUGGCG-------CGUAAGACUGAUAAAGGUUCGAUUGGGU------------- -----------...........(((........((((....))))...((((..(((((((((....))))...(((((((....-------.....)))))))..)))))..)))))))------------- ( -19.20, z-score = -0.74, R) >consensus ___________AAAA_AAAAAAA_______AAAAUGGAUGUUGUAAAUUACUGAAAUCUAAUGGAAACGUUGACGUCAGUUGGUCUCGUGAUGAUAGCUCAAGUCCGCUGAUAAGCCGCACUAUCAACUGAU_ .................................................(((((..((.((((....))))))..)))))..................................................... ( -7.80 = -8.31 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:32 2011