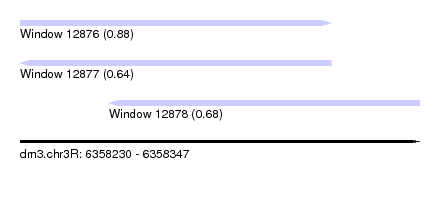
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,358,230 – 6,358,347 |
| Length | 117 |
| Max. P | 0.878602 |

| Location | 6,358,230 – 6,358,321 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Shannon entropy | 0.44473 |
| G+C content | 0.44798 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -10.26 |
| Energy contribution | -10.29 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6358230 91 + 27905053 -----------------------GAGAAGCCGAGAAGACCGAGGCU----GGAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------....((((..(.....)..))))----((......(((((...(((.....)))....)))))...............(((....))).)).... ( -17.40, z-score = -1.60, R) >droSim1.chr3R 12495063 91 - 27517382 -----------------------GAGAAGCCGAGAAGACCGAGGCU----GCAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------....((((..(.....)..))))----((.((...(((((...(((.....)))....))))).)).)).........(((....)))....... ( -17.10, z-score = -1.71, R) >droSec1.super_0 5537071 91 - 21120651 -----------------------GAGAAGCCGAGAAGACCGAGGCU----GGAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------....((((..(.....)..))))----((......(((((...(((.....)))....)))))...............(((....))).)).... ( -17.40, z-score = -1.60, R) >droYak2.chr3R 10387041 91 + 28832112 -----------------------GAGAAGCCGAGAAGACCGAGGCU----GGAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------....((((..(.....)..))))----((......(((((...(((.....)))....)))))...............(((....))).)).... ( -17.40, z-score = -1.60, R) >droEre2.scaffold_4770 2495315 91 - 17746568 -----------------------GAGAAGCCGAGAAGACCGAGGCU----GGAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------....((((..(.....)..))))----((......(((((...(((.....)))....)))))...............(((....))).)).... ( -17.40, z-score = -1.60, R) >droAna3.scaffold_13340 22954890 79 - 23697760 -----------------------------------GAGAGAAGGCC----GGAGAACUUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACGU -----------------------------------...........----((.((..((((((...(((.....)))....))))))))............(((....))).)).... ( -12.60, z-score = -0.67, R) >dp4.chr2 23685928 117 - 30794189 GACUCAGACUGAGACUGAAUCUGAAUCUGAGGCUGAGACCCAGGUCUGGGCCCGAACAUAUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- ..(((.....)))(((((((.((...((((((((.((((....)))).))))......(((((...(((.....)))....))))).))))...)))))))))..............- ( -34.10, z-score = -3.55, R) >droPer1.super_0 10861147 111 - 11822988 ------GACUCAGACUGAGACUGAAUCUGAGGCUGAGACCCAGGUCUGGGCCCGAACAUAUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- ------..(((.....)))(((((((((((((((.((((....)))).))))......(((((...(((.....)))....))))).)))).....)))))))..............- ( -32.40, z-score = -3.62, R) >droWil1.scaffold_181108 146967 76 + 4707319 --------------------------------------CUGAAGC---GACCCGUAGAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- --------------------------------------((((.((---(...)))...(((((...(((.....)))....))))).))))..........(((....)))......- ( -12.20, z-score = -1.34, R) >droVir3.scaffold_13047 1326992 89 + 19223366 -----------------------AAAACGUAGACAGGGCCCAGGCC--AA---GAACAUGUCCAAAGGCGAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- -----------------------.....((.((...(((....)))--..---.....(((((...((((...))))....))))).)).)).........(((....)))......- ( -17.50, z-score = -1.74, R) >droGri2.scaffold_14906 9249419 92 - 14172833 -----------------------GAAACGAAGACAGAGCCCCGUCC--AAGAAGAACAUGUCCAAAGGCGAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- -----------------------.....((.(((........))).--..........(((((...((((...))))....))))).))............(((....)))......- ( -12.20, z-score = -1.17, R) >droMoj3.scaffold_6540 5397659 90 + 34148556 -----------------------AAAACGUAGACAGAGCCCAGGCC--AA--AGAACAUGUCCAAAGGCGAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG- -----------------------.....((.((..(.((....)))--..--......(((((...((((...))))....))))).)).)).........(((....)))......- ( -13.90, z-score = -1.50, R) >consensus _______________________GAGAAGCCGAGAAGACCGAGGCC____GGAGAACAUGUCCAAAGGCAAACCGCCAAAAGGAUAAUCAGCAACAAUUUAGUAUUAUUACACCACG_ ..........................................((..............(((((...(((.....)))....)))))...............(((....))).)).... (-10.26 = -10.29 + 0.03)
| Location | 6,358,230 – 6,358,321 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Shannon entropy | 0.44473 |
| G+C content | 0.44798 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -14.43 |
| Energy contribution | -14.11 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
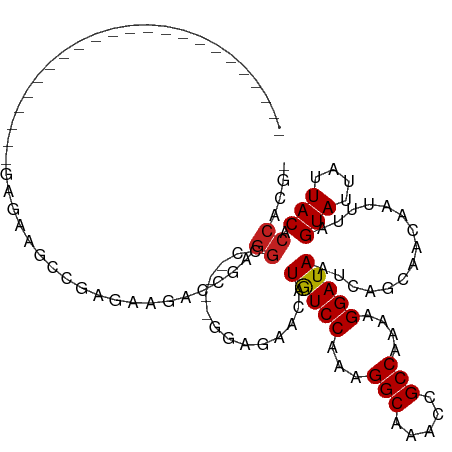
>dm3.chr3R 6358230 91 - 27905053 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUCC----AGCCUCGGUCUUCUCGGCUUCUC----------------------- ...((((((....)))))).......(((((....((....(((((....))..((((.......)))----))))..)).....))))).....----------------------- ( -19.70, z-score = -0.83, R) >droSim1.chr3R 12495063 91 + 27517382 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUGC----AGCCUCGGUCUUCUCGGCUUCUC----------------------- (((((..((((((((.....))))))))......(((....(((.....)))...))))))))...(.----((((..((....)).)))).)..----------------------- ( -19.90, z-score = -0.64, R) >droSec1.super_0 5537071 91 + 21120651 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUCC----AGCCUCGGUCUUCUCGGCUUCUC----------------------- ...((((((....)))))).......(((((....((....(((((....))..((((.......)))----))))..)).....))))).....----------------------- ( -19.70, z-score = -0.83, R) >droYak2.chr3R 10387041 91 - 28832112 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUCC----AGCCUCGGUCUUCUCGGCUUCUC----------------------- ...((((((....)))))).......(((((....((....(((((....))..((((.......)))----))))..)).....))))).....----------------------- ( -19.70, z-score = -0.83, R) >droEre2.scaffold_4770 2495315 91 + 17746568 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUCC----AGCCUCGGUCUUCUCGGCUUCUC----------------------- ...((((((....)))))).......(((((....((....(((((....))..((((.......)))----))))..)).....))))).....----------------------- ( -19.70, z-score = -0.83, R) >droAna3.scaffold_13340 22954890 79 + 23697760 ACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAAGUUCUCC----GGCCUUCUCUC----------------------------------- ....(((((((((((.....))))))))......(((....(((.....)))...)))..........----.))).......----------------------------------- ( -14.80, z-score = -0.25, R) >dp4.chr2 23685928 117 + 30794189 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGAUAUGUUCGGGCCCAGACCUGGGUCUCAGCCUCAGAUUCAGAUUCAGUCUCAGUCUGAGUC -..((((((....)))))).......(((((.(((((....(((.....)))...)))))......((((((....)))))))))))((((((((.((((...)))).)))))))).. ( -40.30, z-score = -3.16, R) >droPer1.super_0 10861147 111 + 11822988 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGAUAUGUUCGGGCCCAGACCUGGGUCUCAGCCUCAGAUUCAGUCUCAGUCUGAGUC------ -..((((((....))))))...((..(((((.(((((....(((.....)))...)))))......((((((....)))))))))))..))(((((((.......)))))))------ ( -36.70, z-score = -2.66, R) >droWil1.scaffold_181108 146967 76 - 4707319 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUCUACGGGUC---GCUUCAG-------------------------------------- -(((((.((((((((.....))))))))......(((....(((.....)))...)))...)))))....---.......-------------------------------------- ( -16.90, z-score = -0.74, R) >droVir3.scaffold_13047 1326992 89 - 19223366 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUC---UU--GGCCUGGGCCCUGUCUACGUUUU----------------------- -(((((.((((((((.....))))))))......(((....((((...))))...))).......---..--(((....)))....)))))....----------------------- ( -21.80, z-score = -0.90, R) >droGri2.scaffold_14906 9249419 92 + 14172833 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUCUUCUU--GGACGGGGCUCUGUCUUCGUUUC----------------------- -((((..((((((((.....))))))))......(((....((((...))))...)))))))........--((((((....)))))).......----------------------- ( -21.30, z-score = -1.18, R) >droMoj3.scaffold_6540 5397659 90 - 34148556 -CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUCU--UU--GGCCUGGGCUCUGUCUACGUUUU----------------------- -((((..((((((((.....)))))))).............(((((...((((..((.((......--.)--).)).)))).)))))))))....----------------------- ( -20.60, z-score = -1.01, R) >consensus _CGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUCUCC____GGCCUCGGUCUUCUCGGCUUCUC_______________________ ....(((((((((((.....))))))))......(((....((((...))))...)))...............))).......................................... (-14.43 = -14.11 + -0.32)

| Location | 6,358,256 – 6,358,347 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Shannon entropy | 0.31052 |
| G+C content | 0.42711 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -16.87 |
| Energy contribution | -16.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6358256 91 - 27905053 -------------CCCAACCCUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC------------ -------------...........(((((.....)))))(((((..((((((((.....))))))))......(((....(((.....)))...))))))))..------------ ( -20.10, z-score = -1.13, R) >droSim1.chr3R 12495089 91 + 27517382 -------------CCCAACCCUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC------------ -------------...........(((((.....)))))(((((..((((((((.....))))))))......(((....(((.....)))...))))))))..------------ ( -20.10, z-score = -1.13, R) >droSec1.super_0 5537097 91 + 21120651 -------------CCCAACCCUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC------------ -------------...........(((((.....)))))(((((..((((((((.....))))))))......(((....(((.....)))...))))))))..------------ ( -20.10, z-score = -1.13, R) >droYak2.chr3R 10387067 91 - 28832112 -------------CCCAACCCUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC------------ -------------...........(((((.....)))))(((((..((((((((.....))))))))......(((....(((.....)))...))))))))..------------ ( -20.10, z-score = -1.13, R) >droEre2.scaffold_4770 2495341 91 + 17746568 -------------CCCAACCCUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC------------ -------------...........(((((.....)))))(((((..((((((((.....))))))))......(((....(((.....)))...))))))))..------------ ( -20.10, z-score = -1.13, R) >droAna3.scaffold_13340 22954904 98 + 23697760 ------CCCCAAGUCCCCGCUUUUUUCGAUGGCUUCGAAACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAAGUUC------------ ------......((((((((...((((((.....)))))).)))).((((((((.....)))))))).............(((.....)))...))))......------------ ( -23.10, z-score = -1.04, R) >dp4.chr2 23685969 102 + 30794189 -------------UUGGGUGCUUCUUCGAUGGCUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGAUAUGUUCGGGCCCAGACCU -------------(((((((((........))).(((-((((((..((((((((.....))))))))......(((....(((.....)))...)))))))))))))))))).... ( -31.60, z-score = -2.27, R) >droPer1.super_0 10861182 102 + 11822988 -------------CUGGGUGCUUCUUCGAUGGCUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGAUAUGUUCGGGCCCAGACCU -------------(((((((((........))).(((-((((((..((((((((.....))))))))......(((....(((.....)))...)))))))))))))))))).... ( -33.90, z-score = -3.08, R) >droWil1.scaffold_181108 146979 103 - 4707319 CCCCCCUUCUUUUGGGGGGCUUCUUUCGAUGGCUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUCUAC------------ ((((((.......))))))........((((((..((-(...((((((....))))))..)))..))......(((....(((.....)))...)))))))...------------ ( -30.90, z-score = -2.47, R) >droVir3.scaffold_13047 1327017 85 - 19223366 ------------------UUAGACUUCGAUGGCUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUC------------ ------------------..................(-((((((..((((((((.....))))))))......(((....((((...))))...))))))))))------------ ( -20.90, z-score = -1.59, R) >droGri2.scaffold_14906 9249447 86 + 14172833 -----------------UUUAGACUUCGAUGGGUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUC------------ -----------------....((((......)))).(-((((((..((((((((.....))))))))......(((....((((...))))...))))))))))------------ ( -21.60, z-score = -1.64, R) >droMoj3.scaffold_6540 5397685 85 - 34148556 ------------------UUAGACUUCGAUGGCUUCG-AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUCGCCUUUGGACAUGUUC------------ ------------------..................(-((((((..((((((((.....))))))))......(((....((((...))))...))))))))))------------ ( -20.90, z-score = -1.59, R) >consensus _____________CCCAACCCUUUUUCGAUGGCUUCG_AACGUGGUGUAAUAAUACUAAAUUGUUGCUGAUUAUCCUUUUGGCGGUUUGCCUUUGGACAUGUUC____________ ..........................(((.....))).((((((..((((((((.....))))))))......(((....((((...))))...)))))))))............. (-16.87 = -16.79 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:31 2011