| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,355,930 – 6,356,010 |
| Length | 80 |
| Max. P | 0.528709 |

| Location | 6,355,930 – 6,356,010 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Shannon entropy | 0.63940 |
| G+C content | 0.40092 |
| Mean single sequence MFE | -14.49 |
| Consensus MFE | -7.30 |
| Energy contribution | -7.03 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
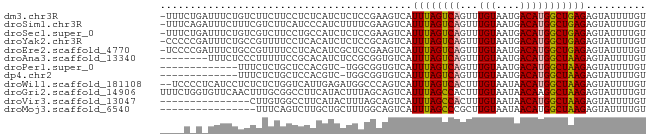
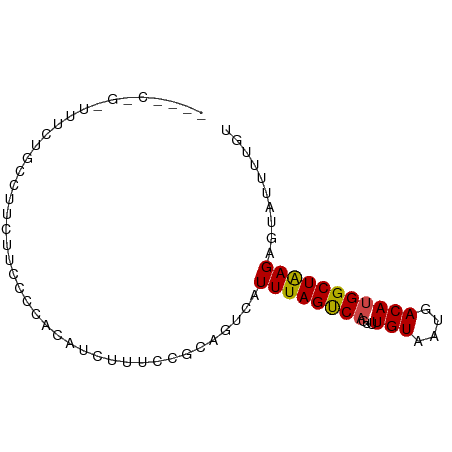
>dm3.chr3R 6355930 80 + 27905053 -UUUCUGAUUUCUGUCUUCUUCCUCUCAUCUCUCCGAAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUGAGAGUAUUUUGU -.....(((....)))......(((((.((.....))((((((...((((........)))))))))))))))........ ( -14.30, z-score = -0.35, R) >droSim1.chr3R 12492761 80 - 27517382 -UUUCAGAUUUCUUUCGUCUUCAUCCCAUCUUUUCGAAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUGAGAGUAUUUUGU -...((((...((((((.((((.............)))).)....((((((((......))).))))))))))...)))). ( -15.52, z-score = -1.44, R) >droSec1.super_0 5534808 80 - 21120651 -UUUCUGAUUUCUGUCGUCUUCCUGCCAUCUCUCCGAAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUGAGAGUAUUUUGU -.....(((....))).............(((((...((((((...((((........)))))))))))))))........ ( -14.50, z-score = -0.25, R) >droYak2.chr3R 10384751 80 + 28832112 -CCCCCGAUUUCUGCCGUUUUCCUCACAUCUCUCCGCAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUGAGAGUAUUUUGU -........................(((.(((((.((.(((((((((.....))).))))))...)).))))).....))) ( -14.50, z-score = -0.86, R) >droEre2.scaffold_4770 2493044 80 - 17746568 -UCCCCGAUUUCUGCCGUUUUCCUCACAUCGCUCCGAAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUGAGAGUAUUUUGU -.......((((.(((((..........(((...))).(((((((((.....))).))))))))))).))))......... ( -14.20, z-score = -0.32, R) >droAna3.scaffold_13340 22952772 73 - 23697760 --------UUUCUCCCUUUUUCCGCACAUCUCCGCGGUGUCAUUUAGUCAGUUUGUAAUGACAUGGCUAAGAGUAUUUUGU --------...(((.......((((........))))......((((((((((......))).))))))))))........ ( -15.40, z-score = -1.87, R) >droPer1.super_0 10858930 67 - 11822988 -------------UUUCUCUGCUCCACGUC-UGGCGGUGUCAUUUAGUCAGUUUGUAAUGACAUGGCUAAGAGUAUUUUGU -------------......(((((......-((((.(((((((((((.....))).)))))))).)))).)))))...... ( -17.10, z-score = -1.75, R) >dp4.chr2 23683739 67 - 30794189 -------------UUUCUCUGCUCCACGUC-UGGCGGUGUCAUUUAGUCAGUUUGUAAUGACAUGGCUAAGAGUAUUUUGU -------------......(((((......-((((.(((((((((((.....))).)))))))).)))).)))))...... ( -17.10, z-score = -1.75, R) >droWil1.scaffold_181108 144766 79 + 4707319 --UCCCCUCAUCCUCUCUCUGGUCAUUGAGAUGGCCCAGUCAUUUAGUCACUUUGUAAUAACAUGGCUAAGAGUAUUUUGU --....(((.........((((((((....)))).))))....(((((((...(((....)))))))))))))........ ( -15.10, z-score = -0.32, R) >droGri2.scaffold_14906 9247155 81 - 14172833 UUUCUGGUGUUCAACUUUGCGGCCUUCAUACUUUAGCAGUCAUUUAGCCACUUUGUAAUAACAAGGCUAAGAGUAUUUUGU .....(((((........)).)))...((((((..........((((((...((((....))))))))))))))))..... ( -11.70, z-score = 1.11, R) >droVir3.scaffold_13047 1325023 66 + 19223366 ---------------CUUGUGGCCUUCAUACUUUAGCAGUCAUUUAGCCACUUUGUAAUAACAUGGCUAAGAGUAUUUUGU ---------------..((((.....)))).....((((.(.((((((((...(((....))))))))))).)....)))) ( -10.30, z-score = 0.56, R) >droMoj3.scaffold_6540 5395813 65 + 34148556 ----------------UUUCAGUCUUGCUGCUUUGGCAGUCAUUUAGCCCGCUUGUAAUAACAUGGCUAAGAGUAUUUUGU ----------------..........(((((....)))))..(((((((....(((....))).))))))).......... ( -14.20, z-score = -0.37, R) >consensus ____C_G_UUUCUGCCUUCUUCCCCACAUCUUUCCGCAGUCAUUUAGUCAGUUUGUAAUGACAUGGCUAAGAGUAUUUUGU ..........................................((((((((...(((....))))))))))).......... ( -7.30 = -7.03 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:28 2011