| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,354,456 – 6,354,547 |
| Length | 91 |
| Max. P | 0.995776 |

| Location | 6,354,456 – 6,354,547 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.42783 |
| G+C content | 0.45663 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -14.73 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
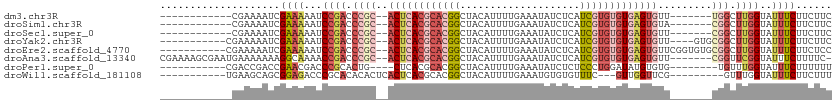
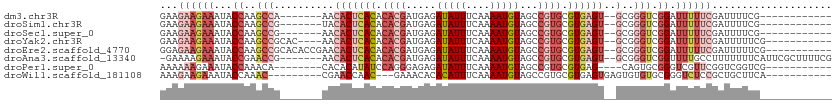
>dm3.chr3R 6354456 91 + 27905053 ------------CGAAAAUCGAAAAAUCCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUU-------UGGCUUGGUAUUUCUUCUUC ------------........((((...((((.((((--((((((((((((........(((......))))))))))))))))).-------.)).))))..))))...... ( -29.90, z-score = -4.28, R) >droSim1.chr3R 12491329 91 - 27517382 ------------CGAAAAUCGAAAAAUCCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUA-------CGGCUUGGUAUUUCUUCUUC ------------........((((...((((.((((--((((((((((((........(((......))))))))))))))))..-------))).))))..))))...... ( -30.30, z-score = -4.33, R) >droSec1.super_0 5533384 91 - 21120651 ------------CGAAAAUCGAAAAAUCCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUU-------CGGCUUGGUAUUUCUUCUUC ------------........((((...((((.((((--((((((((((((........(((......))))))))))))))))..-------))).))))..))))...... ( -30.30, z-score = -4.32, R) >droYak2.chr3R 10383262 95 + 28832112 -----------CGAAAAAUCGAAAAAUCCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUU----GUGCGGCUUGGUAUUUCUUCUUC -----------.((((.(((((.....(((..(.((--((((((((((((........(((......))))))))))))))))).----)..))).))))).))))...... ( -31.20, z-score = -3.86, R) >droEre2.scaffold_4770 2491576 99 - 17746568 -----------CGAAAAAUCGAAAAAUCCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUUCGGUGUGCGGCUUGGUAUUUCUUCUCC -----------.((((.(((((.....(((((((((--((((((((((((........(((......))))))))))))))))..))).)).))).))))).))))...... ( -32.90, z-score = -3.51, R) >droAna3.scaffold_13340 22951354 102 - 23697760 CGAAAAGCGAAUGAAAAAAAGGCAAAACCGACCCGC--ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUU-------CGGUUCGGUAUUUCUUUUC- ................((((((.((.(((((.((((--((((((((((((........(((......))))))))))))))))..-------))).))))).)))))))).- ( -34.10, z-score = -3.77, R) >droPer1.super_0 10857534 89 - 11822988 -----------CGACCGACCGAACGACCCGCACUG----CUCACGCACGGCUACAUUUUGAAAUAUCUCUCCCUGGAUAUGUGUG--------UGUUUGGUAUUUCUUUUUU -----------.((...((((((((...(((((((----(....)))...............((((((......)))))))))))--------))))))))...))...... ( -19.90, z-score = -1.23, R) >droWil1.scaffold_181108 142719 89 + 4707319 -----------UGAAGCAGCGGAGACCCGCACACACUCACUCACGCACGGCUACAUUUUGAAAUGUGUGUUUC---GUUGGUUCG---------GUUUGGUAUUUCUUCUUU -----------.((((..((((....)))).((.(((.(((.(((...(((((((((....)))))).))).)---)).)))..)---------)).))......))))... ( -19.60, z-score = 0.51, R) >consensus ____________GAAAAAUCGAAAAAUCCGACCCGC__ACUCACGCACGGCUACAUUUUGAAAUAUCUCAUCGUGUGUGAGUGUU_______CGGCUUGGUAUUUCUUCUUC ....................((((...((((.(((...((((((((((((....................))))))))))))..........))).))))..))))...... (-14.73 = -16.60 + 1.86)
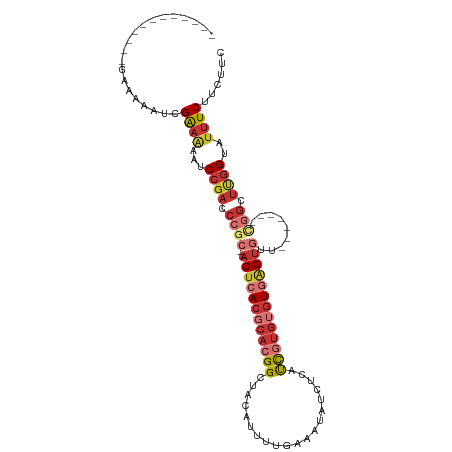
| Location | 6,354,456 – 6,354,547 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.42783 |
| G+C content | 0.45663 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -9.66 |
| Energy contribution | -10.28 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6354456 91 - 27905053 GAAGAAGAAAUACCAAGCCA-------AACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGAUUUUUCGAUUUUCG------------ (((((.((((..((..(((.-------..(((((((.((((..(..(((((....)))))..))))).))))))--)..))).))...))))..))))).------------ ( -28.60, z-score = -3.40, R) >droSim1.chr3R 12491329 91 + 27517382 GAAGAAGAAAUACCAAGCCG-------UACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGAUUUUUCGAUUUUCG------------ (((((.((((..((.(.(((-------..(((((((.((((..(..(((((....)))))..))))).))))))--)))).).))...))))..))))).------------ ( -29.90, z-score = -3.43, R) >droSec1.super_0 5533384 91 + 21120651 GAAGAAGAAAUACCAAGCCG-------AACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGAUUUUUCGAUUUUCG------------ (((((.((((..((.(.(((-------..(((((((.((((..(..(((((....)))))..))))).))))))--)))).).))...))))..))))).------------ ( -29.90, z-score = -3.48, R) >droYak2.chr3R 10383262 95 - 28832112 GAAGAAGAAAUACCAAGCCGCAC----AACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGAUUUUUCGAUUUUUCG----------- (((((.((((..((.(.((((((----.....((((.((((..(..(((((....)))))..))))).))))))--)))).).))...))))...))))).----------- ( -29.00, z-score = -2.92, R) >droEre2.scaffold_4770 2491576 99 + 17746568 GGAGAAGAAAUACCAAGCCGCACACCGAACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGAUUUUUCGAUUUUUCG----------- (((((.((((.......(((.((.(((..(((((((.((((..(..(((((....)))))..))))).))))))--)))))))))...))))...))))).----------- ( -30.60, z-score = -2.72, R) >droAna3.scaffold_13340 22951354 102 + 23697760 -GAAAAGAAAUACCGAACCG-------AACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU--GCGGGUCGGUUUUGCCUUUUUUUCAUUCGCUUUUCG -((((((((..(((((.(((-------..(((((((.((((..(..(((((....)))))..))))).))))))--)))).)))))......))))))))............ ( -33.50, z-score = -3.51, R) >droPer1.super_0 10857534 89 + 11822988 AAAAAAGAAAUACCAAACA--------CACACAUAUCCAGGGAGAGAUAUUUCAAAAUGUAGCCGUGCGUGAG----CAGUGCGGGUCGUUCGGUCGGUCG----------- ......((...(((.((((--------(.(((........((....(((((....)))))..)).(((....)----)))))...)).))).)))...)).----------- ( -18.20, z-score = 0.15, R) >droWil1.scaffold_181108 142719 89 - 4707319 AAAGAAGAAAUACCAAAC---------CGAACCAAC---GAAACACACAUUUCAAAAUGUAGCCGUGCGUGAGUGAGUGUGUGCGGGUCUCCGCUGCUUCA----------- ...((((...........---------.........---...((((((...(((..(((((....)))))...)))))))))((((....))))..)))).----------- ( -21.40, z-score = -0.58, R) >consensus GAAGAAGAAAUACCAAGCCG_______AACACUCACACACGAUGAGAUAUUUCAAAAUGUAGCCGUGCGUGAGU__GCGGGUCGGAUUUUUCGAUUUUUC____________ ...(((((......................((((((.((((.....(((((....)))))...)))).))))))........((((...)))).)))))............. ( -9.66 = -10.28 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:27 2011