
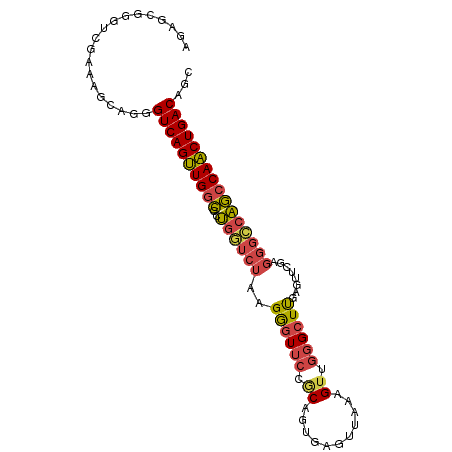
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,333,017 – 6,333,112 |
| Length | 95 |
| Max. P | 0.804739 |

| Location | 6,333,017 – 6,333,112 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Shannon entropy | 0.45050 |
| G+C content | 0.58410 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.50 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.786001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
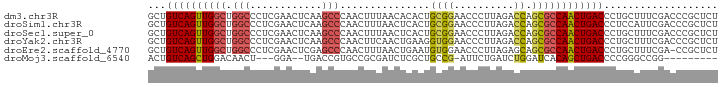
>dm3.chr3R 6333017 95 + 27905053 AGAGCGGGUCGAAAGCAGGGUCAGUUGGCGCUGGUCUAAGGGUUCCGCAGUGUGUUAAAGUUGGGCUUGAGUUCGAGGGCCAGCCAACUGACAGC ...((..(.(....))...((((((((((..((((((..(..(((...(((.((.......)).))).)))..)..)))))))))))))))).)) ( -34.00, z-score = -1.24, R) >droSim1.chr3R 12469990 95 - 27517382 AGAGCGGGUCGAAUGGAGGGUCAGUUGGCGCUGGUCUAAGGGUUCCGCAGUGAGUUAAAGUUGGGCUUGAGUUCGAGGGCCAGCCAACUGACAGC ...................((((((((((..((((((..(..(((......(((((.......))))))))..)..))))))))))))))))... ( -31.90, z-score = -0.77, R) >droSec1.super_0 5512126 95 - 21120651 AGAGCGGGUCGAAAGCAGGGUCAGUUGGCGCUGGUCUAAGGGUUCCGCAGUGAGUUAAAGUUGGGCUUGAGUUCGAGGGCCAGCCAACUGACAGC ...((..(.(....))...((((((((((..((((((..(..(((......(((((.......))))))))..)..)))))))))))))))).)) ( -33.70, z-score = -1.21, R) >droYak2.chr3R 10361192 95 + 28832112 AGAGCGGGUCGAAAGCAGGGUCAGUUGGCGCUGGUCUAAGGGUUCCACCUUCAGUUGAAGUUGGGCUUGAGUUCGAGGGCCAGCCAACUGACAGC ...((..(.(....))...((((((((((..(((((.......((((.((((....)))).))))((((....))))))))))))))))))).)) ( -37.30, z-score = -1.69, R) >droEre2.scaffold_4770 2470239 94 - 17746568 AGAGCGG-UCGAAAGCAGGGUCAGUUGGCGCUGCUCUAAGGGUUCCACAUUCAGUUAAAGUUGGGCUCGAGUUCGAGGGCCAGCCAACUGACAGC ...((..-......))...((((((((((...(((((..((((((.((...........)).)))))).......)))))..))))))))))... ( -32.90, z-score = -1.25, R) >droMoj3.scaffold_6540 5369310 80 + 34148556 ---------CCGGCCCGGGGUCAGCUGUGAUCCAGAUCAGAAU-CGGCAGCGAGAUCGCGGCACGGUCA--UCC---AGUUGUCCAGCUGACAGU ---------(((...))).((((((((((((....))))....-.(((((((((((((.....))))).--)).---.))))))))))))))... ( -28.80, z-score = -0.61, R) >consensus AGAGCGGGUCGAAAGCAGGGUCAGUUGGCGCUGGUCUAAGGGUUCCGCAGUGAGUUAAAGUUGGGCUUGAGUUCGAGGGCCAGCCAACUGACAGC ...................((((((((((..((((((..(..(((......(((((.......))))))))..)..))))))))))))))))... (-23.26 = -24.50 + 1.24)


| Location | 6,333,017 – 6,333,112 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Shannon entropy | 0.45050 |
| G+C content | 0.58410 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6333017 95 - 27905053 GCUGUCAGUUGGCUGGCCCUCGAACUCAAGCCCAACUUUAACACACUGCGGAACCCUUAGACCAGCGCCAACUGACCCUGCUUUCGACCCGCUCU ((.((((((((((.(((............)))...............((((..........)).))))))))))))...)).............. ( -23.60, z-score = -1.33, R) >droSim1.chr3R 12469990 95 + 27517382 GCUGUCAGUUGGCUGGCCCUCGAACUCAAGCCCAACUUUAACUCACUGCGGAACCCUUAGACCAGCGCCAACUGACCCUCCAUUCGACCCGCUCU ...((((((((((.(((............)))...............((((..........)).))))))))))))................... ( -21.80, z-score = -1.19, R) >droSec1.super_0 5512126 95 + 21120651 GCUGUCAGUUGGCUGGCCCUCGAACUCAAGCCCAACUUUAACUCACUGCGGAACCCUUAGACCAGCGCCAACUGACCCUGCUUUCGACCCGCUCU ((.((((((((((.(((............)))...............((((..........)).))))))))))))...)).............. ( -23.60, z-score = -1.30, R) >droYak2.chr3R 10361192 95 - 28832112 GCUGUCAGUUGGCUGGCCCUCGAACUCAAGCCCAACUUCAACUGAAGGUGGAACCCUUAGACCAGCGCCAACUGACCCUGCUUUCGACCCGCUCU ((.(((((((((((((.....(....)....(((.((((....)))).)))..........)))..))))))))))...)).............. ( -27.50, z-score = -1.40, R) >droEre2.scaffold_4770 2470239 94 + 17746568 GCUGUCAGUUGGCUGGCCCUCGAACUCGAGCCCAACUUUAACUGAAUGUGGAACCCUUAGAGCAGCGCCAACUGACCCUGCUUUCGA-CCGCUCU ((.(((((((((((((..((((....)))).))).......(((..((.((...)).))...))).))))))))))...))......-....... ( -28.70, z-score = -2.13, R) >droMoj3.scaffold_6540 5369310 80 - 34148556 ACUGUCAGCUGGACAACU---GGA--UGACCGUGCCGCGAUCUCGCUGCCG-AUUCUGAUCUGGAUCACAGCUGACCCCGGGCCGG--------- ...((((((((((...(.---.((--(.(.((.((.(((....))).))))-....).)))..).)).)))))))).(((...)))--------- ( -27.30, z-score = -0.92, R) >consensus GCUGUCAGUUGGCUGGCCCUCGAACUCAAGCCCAACUUUAACUCACUGCGGAACCCUUAGACCAGCGCCAACUGACCCUGCUUUCGACCCGCUCU ...((((((((((.(((............)))...............((((..........)).))))))))))))................... (-15.90 = -16.10 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:20 2011