
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,324,751 – 6,324,884 |
| Length | 133 |
| Max. P | 0.835568 |

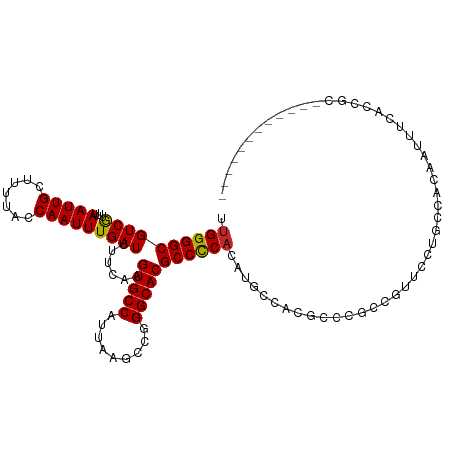
| Location | 6,324,751 – 6,324,854 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.46937 |
| G+C content | 0.52877 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
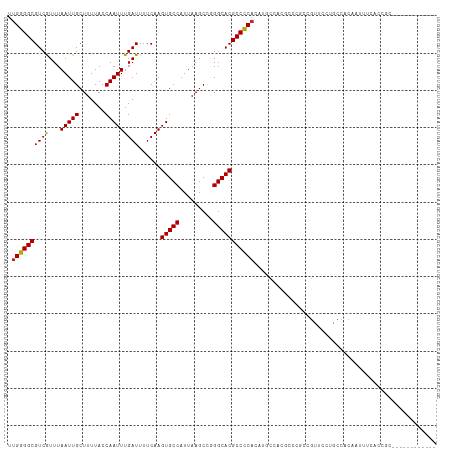
>dm3.chr3R 6324751 103 + 27905053 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGUUCCUGCCACAAUUUCACCGC------------ ...((((((.((......((((......(((((.....)))))......)))).(((....))).....)).)))))).........................------------ ( -23.80, z-score = 0.51, R) >droSim1.chr3R 12461705 103 - 27517382 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGUUCCUGCCACAAUUUCACCGC------------ ...((((((.((......((((......(((((.....)))))......)))).(((....))).....)).)))))).........................------------ ( -23.80, z-score = 0.51, R) >droSec1.super_0 5503920 103 - 21120651 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGUUCCUGCCACAAUUUCACCGC------------ ...((((((.((......((((......(((((.....)))))......)))).(((....))).....)).)))))).........................------------ ( -23.80, z-score = 0.51, R) >droYak2.chr3R 10352633 103 + 28832112 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCCCCGUUUCUGGCGCAAUUUCACCGC------------ .((((((((.((((....((((......(((((.....)))))......)))).)))))))))))).......((((..........))))............------------ ( -27.90, z-score = -0.26, R) >droEre2.scaffold_4770 2461905 103 - 17746568 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCAGCCGUUCCUGCCACAAUUUCACCGC------------ .((((((((.((((....((((......(((((.....)))))......)))).))))))))))))........(.(((......))).).............------------ ( -24.70, z-score = 0.14, R) >dp4.chr2 23651943 109 - 30794189 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCAGGCACGCCCCACAUGCCACAGUAGCAGC-ACC-CCCCCUACCGCUACCCAUCGCCC---- .((((((((.((((....((((......(((((.....)))))......)))).))))))))))))...((....(((((...-...-.........))))).....))..---- ( -28.56, z-score = -2.22, R) >droPer1.super_0 10826369 115 - 11822988 UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCAGGCACGCCCCACAUGCCACAGUAACAGCUACCGCCCCCCACCGCUACCCAUCACCCUCCC ..((((((..(((((((.((........(((((.....))))))).)))))))..((((.(.....).))))..............))))))....................... ( -24.90, z-score = -1.33, R) >droWil1.scaffold_181108 106505 87 + 4707319 UUGGGGCGUCAUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCAGGCACGCCCCA-AGCCCAUACCCACAAGUUCCC--------------------------- (((((((((((...(((((.......)))))))))......(((((.........)))))))))))-)....................--------------------------- ( -23.40, z-score = -2.78, R) >droGri2.scaffold_14906 9206548 112 - 14172833 UUGGGGCGUCAUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCUGGCACGCCUC-CGUUCCCCCGCACCCGCCUCUGCCACCAGACGCCACACCUCCUUGCC-- .((.((((((........((((......(((((.....)))))......)))).(((((((....-)))......((....))....))))...)))))).))..........-- ( -25.30, z-score = -0.96, R) >droMoj3.scaffold_6540 5358596 90 + 34148556 UUGGGGCGUCAUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCUCGGCACGCCCCACCGCCCAACCACCCAGCCACAGCC------------------------- .((((((((((...(((((.......)))))))))......(((((.........)))))))))))........................------------------------- ( -22.40, z-score = -1.69, R) >consensus UUGGGGCGUCGUUUAAUUGCUUUUACCAAUUUGAUUUUCAAGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGUUCCUGCCACAAUUUCACCGC____________ .((((((((((...(((((.......)))))))))......(((((.........)))))))))))................................................. (-21.46 = -21.26 + -0.20)

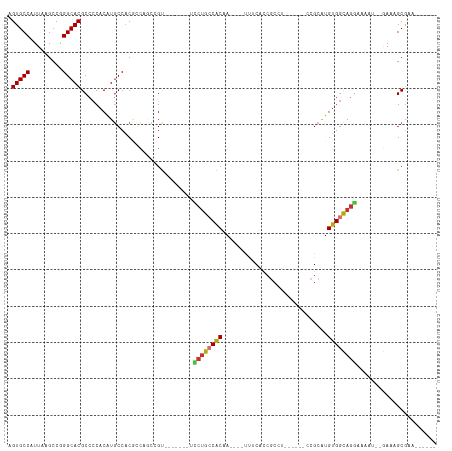
| Location | 6,324,791 – 6,324,884 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Shannon entropy | 0.42243 |
| G+C content | 0.60550 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 6324791 93 + 27905053 AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGU-------UCCUGCCACAA----UUUCACCGCCU------CCGCAUGUGGCAGGAAAAU--GAAAGCGAA------ .((..((((..(.(((((..((.......))...))))).).(-------((((((((((.----...........------.....))))))))))))))--)...))...------ ( -30.03, z-score = -1.35, R) >droSim1.chr3R 12461745 93 - 27517382 AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGU-------UCCUGCCACAA----UUUCACCGCCU------CCGCAUGUGGCAGGAAAAU--GAAAGCGAA------ .((..((((..(.(((((..((.......))...))))).).(-------((((((((((.----...........------.....))))))))))))))--)...))...------ ( -30.03, z-score = -1.35, R) >droSec1.super_0 5503960 93 - 21120651 AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCGCCGU-------UCCUGCCACAA----UUUCACCGCCU------CCGCAUGUGGCAAGAAAAU--GAAAGCGAA------ .((..((((..(.(((((..((.......))...))))).).(-------((.(((((((.----...........------.....))))))).))))))--)...))...------ ( -23.53, z-score = 0.19, R) >droYak2.chr3R 10352673 93 + 28832112 AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCCCCCGU-------UUCUGGCGCAA----UUUCACCGCCU------CCGCAUGUGGCAGGAAAAU--GAAAGCGAA------ .((((((..((((.((((..((.......))...))))...))-------)).))))))..----(((((...(((------(((....))).)))....)--)))).....------ ( -27.40, z-score = 0.22, R) >droEre2.scaffold_4770 2461945 93 - 17746568 AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCAGCCGU-------UCCUGCCACAA----UUUCACCGCCU------CCGCGUGUGGCAGGAAAAU--GAAAGCGAA------ .(((((.........))))).............(((...(..(-------((((((((((.----......(((..------..))))))))))))))...--)...)))..------ ( -32.41, z-score = -1.81, R) >dp4.chr2 23651983 107 - 30794189 AGUGCCAUUAAGCCAGGCACGCCCCACAUGCCACAGUAGCAGC-ACC-CCCCCUACCGCUA----CC-CAUCGCCC----GCCCGCAUGUGGUAGGAAAAGCGAAAAGCGAAAGCGAA .(((((.........)))))((......((((((((((((...-...-.........))))----).-....((..----....)).)))))))......)).....((....))... ( -30.66, z-score = -2.18, R) >droPer1.super_0 10826409 113 - 11822988 AGUGCCAUUAAGCCAGGCACGCCCCACAUGCCACAGUAACAGCUACCGCCCCCCACCGCUA----CC-CAUCACCCUCCCGCCCGCAUGUGGUAGAAAAAGCGAAAAGCGAAAGCGAA .(((((.........)))))...((((((((...(((....)))...((........))..----..-................))))))))...............((....))... ( -26.80, z-score = -2.11, R) >droGri2.scaffold_14906 9206588 101 - 14172833 AGUGCCAUUAAGCCUGGCACGCCUC-CGUUCCCCCGCACCCGC-------CUCUGCCACCAGACGCCACACCUCCUUGCCGCCCGCAUGUGCCAGAAAGUG---AGAGCAAA------ .((((((.......))))))(((((-(.(((....((((..((-------.((((....)))).))..........(((.....))).))))..))).).)---)).))...------ ( -27.60, z-score = -1.76, R) >consensus AGUGCCAUUAAGCCGGGCACGCCCCACAUGCCACGCCAGCCGU_______UCCUGCCACAA____UUUCACCGCCU______CCGCAUGUGGCAGGAAAAU__GAAAGCGAA______ .(((((.........)))))...............................((((((((.............................))))))))...................... (-17.43 = -17.20 + -0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:18 2011