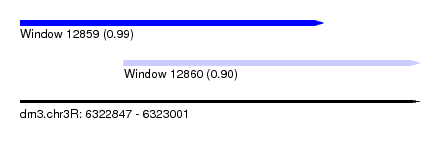
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 6,322,847 – 6,323,001 |
| Length | 154 |
| Max. P | 0.993793 |

| Location | 6,322,847 – 6,322,964 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.94 |
| Shannon entropy | 0.02356 |
| G+C content | 0.37538 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
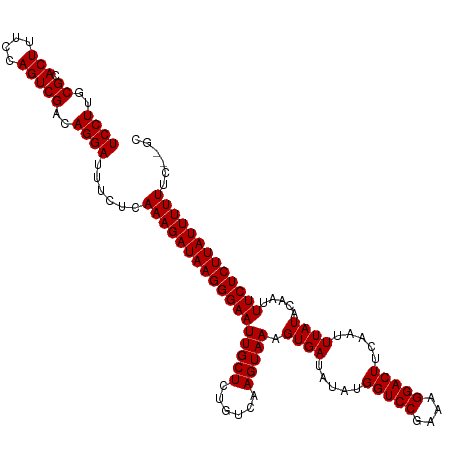
>dm3.chr3R 6322847 117 + 27905053 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.75, R) >droAna3.scaffold_13340 22922237 117 - 23697760 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.75, R) >droEre2.scaffold_4770 2456147 117 - 17746568 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.75, R) >droYak2.chr3R 10350407 117 + 28832112 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.75, R) >droSec1.super_0 5501983 117 - 21120651 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.75, R) >droMoj3.scaffold_6540 5357018 118 + 34148556 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUCC-GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))...-.. ( -30.80, z-score = -2.57, R) >droVir3.scaffold_13047 1288801 118 + 19223366 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUCC-GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))...-.. ( -30.80, z-score = -2.57, R) >droGri2.scaffold_14906 9204389 118 - 14172833 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUCC-GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))...-.. ( -30.80, z-score = -2.57, R) >droWil1.scaffold_181108 104671 119 + 4707319 UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUUUCGC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))...... ( -30.80, z-score = -2.73, R) >dp4.chr2 23648803 117 - 30794189 UCCUUGCGUACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.81, R) >droPer1.super_0 10823250 117 - 11822988 UCCUUGCGUACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))..--.. ( -30.80, z-score = -2.81, R) >consensus UCCUUGCGCACUUUCCAGUCGACAGGAUUUCUCAAAGAUAAGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC__GC ((((..((.(((....)))))..))))......((((((((((((((((((......))))).((((.....(((((....))))).....)))).....)))))))))))))...... (-30.80 = -30.80 + 0.00)

| Location | 6,322,887 – 6,323,001 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Shannon entropy | 0.27035 |
| G+C content | 0.36456 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.11 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
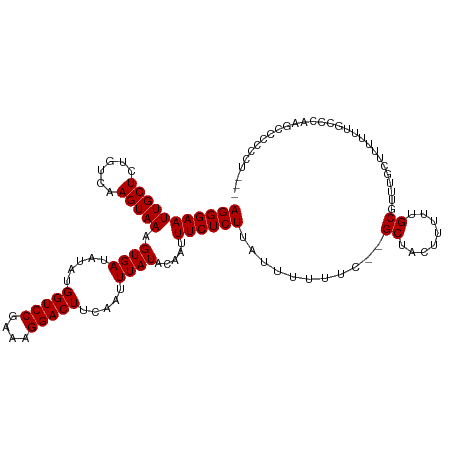
>dm3.chr3R 6322887 114 + 27905053 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUUUUCCCAAGCCCCCCAU-- .(((....(((.((..(((.((((..(.....(((((....)))))............................(--((........))).)..)))).)))..)))))..)))..-- ( -24.90, z-score = -2.11, R) >droAna3.scaffold_13340 22922277 116 - 23697760 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUUUCGCCCCGCCCCUGCCCG .(((....((......(((((((((((.....(((((....))))).....))))...................(--((........)))))))))).....))....)))....... ( -24.40, z-score = -1.29, R) >droEre2.scaffold_4770 2456187 111 - 17746568 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUU--UCCCAAGCCCCCCU--- .(((((..((......(((..((((..((((.(((((....)))))......))))..))))..))).......(--((........)))...))...)--))))..........--- ( -25.90, z-score = -2.48, R) >droYak2.chr3R 10350447 115 + 28832112 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUU-UCCCAAGUCCCCCCCCU .((((.(((.......(((((((((((.....(((((....))))).....))))...................(--((........))))))))))...-...))).))))...... ( -25.72, z-score = -2.45, R) >droSec1.super_0 5502023 113 - 21120651 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUU-UCCCAAGCCCCCCUU-- .(((((..((......(((..((((..((((.(((((....)))))......))))..))))..))).......(--((........)))...))....)-))))...........-- ( -25.20, z-score = -2.06, R) >droMoj3.scaffold_6540 5357058 106 + 34148556 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC-CGCUACUUUUUGCGUUUUUCUGUAUGCUCUC----------- (((((((((((......))))).((((.....(((((....))))).....)))).....)))))).........-(((........))).................----------- ( -20.60, z-score = -1.30, R) >droVir3.scaffold_13047 1288841 106 + 19223366 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC-CGCUACUUUUUGCGUUUUUCUGUAUGCUCUC----------- (((((((((((......))))).((((.....(((((....))))).....)))).....)))))).........-(((........))).................----------- ( -20.60, z-score = -1.30, R) >droGri2.scaffold_14906 9204429 103 - 14172833 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC-CGCUACUUUUUGCGUUUUUCUGUAUGCU-------------- (((((((((((......))))).((((.....(((((....))))).....)))).....)))))).........-(((........)))..............-------------- ( -20.60, z-score = -1.32, R) >droWil1.scaffold_181108 104711 117 + 4707319 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUUUCGCUACUUUUUGCGUUUUUCUGUUUGCUUUCUCGCUCUCUC- .(((((..((......(((..((((..((((.(((((....)))))......))))..))))..))).........(((........)))...........)).)))))........- ( -22.50, z-score = -1.74, R) >dp4.chr2 23648843 116 - 30794189 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUUUCACCAUCACCAUCUCCU .((((.(((((......))))).(((((....(((((....)))))............................(--((........)))...............)))))....)))) ( -22.30, z-score = -1.71, R) >droPer1.super_0 10823290 116 - 11822988 AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC--GCUACUUUUUGCGUUUGCUUUUUUCACCAUCACCAUCUCCU .((((.(((((......))))).(((((....(((((....)))))............................(--((........)))...............)))))....)))) ( -22.30, z-score = -1.71, R) >consensus AGGGAAUUGCUCUGUCAAGUAAAGUGAUAUAUGGUCCGAAAGGACUUCAAUUUAUACAAUUUCUCUUAUUUUUUC__GCUACUUUUUGCGUUUGCUUUUUUGCCCAAGCCCCCCU___ (((((((((((......))))).((((.....(((((....))))).....)))).....))))))...........((........))............................. (-19.11 = -19.11 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:03:16 2011