| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,871,638 – 6,871,778 |
| Length | 140 |
| Max. P | 0.839679 |

| Location | 6,871,638 – 6,871,778 |
|---|---|
| Length | 140 |
| Sequences | 7 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.43558 |
| G+C content | 0.29849 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -15.88 |
| Energy contribution | -17.40 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
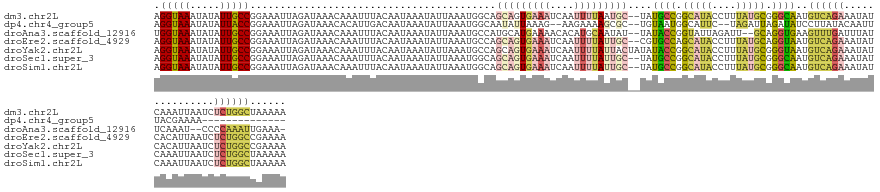
>dm3.chr2L 6871638 140 - 23011544 AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGGCAGCAGUGAAAUCAAUUUUAAUGC--UAUGCCGGCAUACCUUUAUGCGGGCAAUGUCAGAAAUAUCAAAUUAAUCUCUGGCUAAAAA .............((((((.(((((.((((....))))......(((((...((((((((.(((((....))))).)))--).((((.(((((....))))).))))..))))..)))))....))))).))))))...... ( -33.30, z-score = -2.55, R) >dp4.chr4_group5 370005 122 - 2436548 AGGUAAAUAUAUUACCGGAAAUUAGAUAAACACAUUGACAAUAAAUAUUAAAUGGCAAUAUUAAAG--AAGAAAAGCGC--UGUAAUGGCAUUC--UAGAUUAGAUAUCCUUAUACAAUUUACGAAAA-------------- .(((((.....)))))(((.(((.(((.....((((..............))))............--.........((--(.....)))....--...))).))).)))..................-------------- ( -11.94, z-score = 0.86, R) >droAna3.scaffold_12916 8459359 135 - 16180835 UGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGCCAUGCAUGAAAACACAUGCAAUAU--UAUACCGGUAUUAGAUU--GCAGGUGAAGUUUGAUUUAUUCAAAU--CCCCAAAUUGAAA- .(((((.....)))))(((..((..((((((((((((....................((((((......))))))....--..((((.(((......)--)).))))))))))).)))))..)).)--))...........- ( -26.30, z-score = -1.52, R) >droEre2.scaffold_4929 15790397 140 - 26641161 AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGCCAGCAGUGAAAUCAAUUUUAUUGC--CGUGCCAGCAUACCUUUAUGCAGGUAAUGUCAGAAAUAUCACAUUAAUCUCUGGCCGAAAA .............((((((.((((((((.............................(((((((((....)))))))))--..((((.(((((....))))).)))).)))).((....))....)))).))))))...... ( -28.60, z-score = -1.31, R) >droYak2.chr2L 16291731 142 + 22324452 AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGCCAGCAGUGAAAUCAAUUUUAUUACUAUAUACCGGCAUACCUUUAUGCGGGUAAUGUCAGAAAUAUCACAUUAAUCUCUGGCCGAAAA .............((((((.((((((((............................((.(((((((....))))))).))...((((.(((((....))))).)))).)))).((....))....)))).))))))...... ( -26.20, z-score = -0.94, R) >droSec1.super_3 2411578 140 - 7220098 AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGGCAGCAGUGAAAUCAAUUUUAUUGC--UAUGCCGGCAUACCUUUAUGCGGGCAAUGUCAGAAAUAUCAAAUUAAUCUCUGGCUAAAAA .............((((((.(((((.((((....))))......(((((...((((((((((((((....)))))))))--).((((.(((((....))))).))))..))))..)))))....))))).))))))...... ( -37.00, z-score = -3.61, R) >droSim1.chr2L 6672400 140 - 22036055 AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGGCAGCAGUGAAAUCAAUUUUAUUGC--UAUGCCGGCAUACCUUUAUGCGGGCAAUGUCAGAAAUAUCAAAUUAAUCUCUGGCUAAAAA .............((((((.(((((.((((....))))......(((((...((((((((((((((....)))))))))--).((((.(((((....))))).))))..))))..)))))....))))).))))))...... ( -37.00, z-score = -3.61, R) >consensus AGGUAAAUAUAUUGCCGGAAAUUAGAUAAACAAAUUUACAAUAAAUAUUAAAUGGCAGCAGUGAAAUCAAUUUUAUUGC__UAUGCCGGCAUACCUUUAUGCGGGUAAUGUCAGAAAUAUCAAAUUAAUCUCUGGCUAAAAA .(((((.....))))).........................................(((((((((....)))))))))....((((.(((((....))))).))))..((((((...............))))))...... (-15.88 = -17.40 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:27 2011